A Distinct Transcriptional Program in Human CAR T Cells Bearing the 4-1BB Signaling Domain Revealed by scRNA-Seq
- PMID: 32755564
- PMCID: PMC7704462
- DOI: "VSports最新版本" 10.1016/j.ymthe.2020.07.023
A Distinct Transcriptional Program in Human CAR T Cells Bearing the 4-1BB Signaling Domain Revealed by scRNA-Seq
Abstract
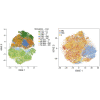
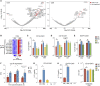
T cells engineered to express chimeric antigen receptors (CARs) targeting CD19 have produced impressive outcomes for the treatment of B cell malignancies, but different products vary in kinetics, persistence, and toxicity profiles based on the co-stimulatory domains included in the CAR. In this study, we performed transcriptional profiling of bulk CAR T cell populations and single cells to characterize the transcriptional states of human T cells transduced with CD3ζ, 4-1BB-CD3ζ (BBζ), or CD28-CD3ζ (28ζ) co-stimulatory domains at rest and after activation by triggering their CAR or their endogenous T cell receptor (TCR). We identified a transcriptional signature common across CARs with the CD3ζ signaling domain, as well as a distinct program associated with the 4-1BB co-stimulatory domain at rest and after activation. CAR T cells bearing BBζ had increased expression of human leukocyte antigen (HLA) class II genes, ENPP2, and interleukin (IL)-21 axis genes, and decreased PD1 compared to 28ζ CAR T cells. Similar to previous studies, we also found BBζ CAR CD8 T cells to be enriched in a central memory cell phenotype and fatty acid metabolism genes. Our data uncovered transcriptional signatures related to costimulatory domains and demonstrated that signaling domains included in CARs uniquely shape the transcriptional programs of T cells VSports手机版. .
Keywords: CAR T cells; T cell activation; adoptive cellular therapy; single-cell RNA sequencing; transcriptional profiling. V体育安卓版.
Copyright © 2020 The American Society of Gene and Cell Therapy. Published by Elsevier Inc. All rights reserved V体育ios版. .
Figures




References (VSports在线直播)
-
- Kalos M., Levine B.L., Porter D.L., Katz S., Grupp S.A., Bagg A., June C.H. T cells with chimeric antigen receptors have potent antitumor effects and can establish memory in patients with advanced leukemia. Sci. Transl. Med. 2011;3:95ra73. - "V体育平台登录" PMC - PubMed
-
- Brentjens R.J., Davila M.L., Riviere I., Park J., Wang X., Cowell L.G., Bartido S., Stefanski J., Taylor C., Olszewska M. CD19-targeted T cells rapidly induce molecular remissions in adults with chemotherapy-refractory acute lymphoblastic leukemia. Sci. Transl. Med. 2013;5:177ra38. - PMC (VSports在线直播) - PubMed
-
- Kochenderfer J.N., Dudley M.E., Kassim S.H., Somerville R.P., Carpenter R.O., Stetler-Stevenson M., Yang J.C., Phan G.Q., Hughes M.S., Sherry R.M. Chemotherapy-refractory diffuse large B-cell lymphoma and indolent B-cell malignancies can be effectively treated with autologous T cells expressing an anti-CD19 chimeric antigen receptor. J. Clin. Oncol. 2015;33:540–549. - PMC - PubMed
-
- Hamza T., Barnett J.B., Li B. Interleukin 12 a key immunoregulatory cytokine in infection applications. Int. J. Mol. Sci. 2010;11:789–806. - PMC (VSports注册入口) - PubMed
"VSports注册入口" Publication types
- Actions (VSports最新版本)
- "VSports app下载" Actions
"VSports在线直播" MeSH terms
- "VSports手机版" Actions
- Actions (VSports在线直播)
- "VSports" Actions
- V体育ios版 - Actions
- V体育官网 - Actions
- VSports在线直播 - Actions
V体育官网入口 - Substances
- "V体育官网入口" Actions
"VSports注册入口" Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
"VSports" Research Materials

