The chromatin landscape at the HIV-1 provirus integration site determines viral expression (VSports在线直播)
- PMID: 32597987
- PMCID: PMC7641320
- DOI: 10.1093/nar/gkaa536
The chromatin landscape at the HIV-1 provirus integration site determines viral expression
Abstract (V体育ios版)
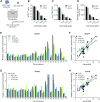
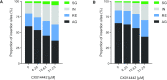
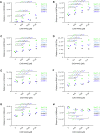
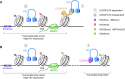
HIV-1 persists lifelong in memory cells of the immune system as latent provirus that rebounds upon treatment interruption. Therefore, the latent reservoir is the main target for an HIV cure. Here, we studied the direct link between integration site and transcription using LEDGINs and Barcoded HIV-ensembles (B-HIVE). LEDGINs are antivirals that inhibit the interaction between HIV-1 integrase and the chromatin-tethering factor LEDGF/p75. They were used as a tool to retarget integration, while the effect on HIV expression was measured with B-HIVE. B-HIVE tracks insert-specific HIV expression by tagging a unique barcode in the HIV genome. We confirmed that LEDGINs retarget integration out of gene-dense and actively transcribed regions. The distance to H3K36me3, the marker recognized by LEDGF/p75, clearly increased. LEDGIN treatment reduced viral RNA expression and increased the proportion of silent provirus. Finally, silent proviruses obtained after LEDGIN treatment were located further away from epigenetic marks associated with active transcription VSports手机版. Interestingly, proximity to enhancers stimulated transcription irrespective of LEDGIN treatment, while the distance to H3K36me3 only changed after treatment with LEDGINs. The fact that proximity to these markers are associated with RNA expression support the direct link between provirus integration site and viral expression. .
© The Author(s) 2020. Published by Oxford University Press on behalf of Nucleic Acids Research V体育安卓版. .
Figures



References (V体育2025版)
-
- Finzi D., Hermankova M., Pierson T., Carruth L.M., Buck C., Chaisson R.E., Quinn T.C., Chadwick K., Margolick J., Brookmeyer R. et al. .. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997; 278:1295–1300. - PubMed
-
- Finzi D., Blankson J., Siliciano J.D., Margolick J.B., Chadwick K., Pierson T., Smith K., Lisziewicz J., Lori F., Flexner C. et al. .. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat. Med. 1999; 5:512. - PubMed
-
- Chomont N., El-Far M., Ancuta P., Trautmann L., Procopio F.A., Yassine-Diab B., Boucher G., Boulassel M.-R., Ghattas G., Brenchley J.M. et al. .. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat. Med. 2009; 15:893. - PMC (VSports app下载) - PubMed
-
- Chun T.-W., Carruth L., Finzi D., Shen X., DiGiuseppe J.A., Taylor H., Hermankova M., Chadwick K., Margolick J., Quinn T.C. et al. .. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature. 1997; 387:183–188. - PubMed
V体育官网入口 - Publication types
- Actions (VSports最新版本)
MeSH terms
- "VSports最新版本" Actions
- Actions (VSports手机版)
- "VSports" Actions
- "V体育官网入口" Actions
- Actions (VSports注册入口)
- "VSports app下载" Actions
- "VSports在线直播" Actions
- Actions (V体育官网)
"V体育平台登录" Substances
- Actions (V体育ios版)
- "VSports" Actions
- V体育ios版 - Actions
- Actions (V体育2025版)
LinkOut - more resources
"V体育平台登录" Full Text Sources
Research Materials

