Single-cell transcriptomics identifies an effectorness gradient shaping the response of CD4+ T cells to cytokines (V体育2025版)
- PMID: 32286271
- PMCID: PMC7156481
- DOI: 10.1038/s41467-020-15543-y
Single-cell transcriptomics identifies an effectorness gradient shaping the response of CD4+ T cells to cytokines
Abstract
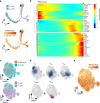
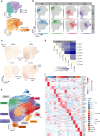
Naïve CD4+ T cells coordinate the immune response by acquiring an effector phenotype in response to cytokines. However, the cytokine responses in memory T cells remain largely understudied. Here we use quantitative proteomics, bulk RNA-seq, and single-cell RNA-seq of over 40,000 human naïve and memory CD4+ T cells to show that responses to cytokines differ substantially between these cell types. Memory T cells are unable to differentiate into the Th2 phenotype, and acquire a Th17-like phenotype in response to iTreg polarization. Single-cell analyses show that T cells constitute a transcriptional continuum that progresses from naïve to central and effector memory T cells, forming an effectorness gradient accompanied by an increase in the expression of chemokines and cytokines VSports手机版. Finally, we show that T cell activation and cytokine responses are influenced by the effectorness gradient. Our results illustrate the heterogeneity of T cell responses, furthering our understanding of inflammation. .
Conflict of interest statement
All authors declare no competing interests.
V体育2025版 - Figures




References
-
- Mosmann TR, Cherwinski H, Bond MW, Giedlin MA, Coffman RL. Two types of murine helper T cell clone. I. Definition according to profiles of lymphokine activities and secreted proteins. J. Immunol. 1986;136:2348–2357. - PubMed
-
- Le Gros G, Ben-Sasson SZ, Seder R, Finkelman FD, Paul WE. Generation of interleukin 4 (IL-4)-producing cells in vivo and in vitro: IL-2 and IL-4 are required for in vitro generation of IL-4-producing cells. J. Immunol. 2008;181:2943–2951. - PubMed
Publication types
MeSH terms
- "V体育官网" Actions
- V体育官网入口 - Actions
- Actions (VSports)
- "V体育2025版" Actions
- Actions (V体育2025版)
- "VSports在线直播" Actions
- "V体育官网" Actions
- "VSports注册入口" Actions
- Actions (VSports注册入口)
Substances
- VSports - Actions
- "VSports注册入口" Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

