The ubiquitin ligase FBXW7 targets the centriolar assembly protein HsSAS-6 for degradation and thereby regulates centriole duplication
- PMID: 32086376
- PMCID: PMC7135989
- DOI: 10.1074/jbc.AC119.012178
The ubiquitin ligase FBXW7 targets the centriolar assembly protein HsSAS-6 for degradation and thereby regulates centriole duplication
Abstract (VSports最新版本)
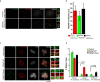
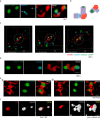
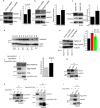
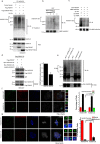
Formation of a single new centriole from a pre-existing centriole is strictly controlled to maintain correct centrosome number and spindle polarity in cells VSports手机版. However, the mechanisms that govern this process are incompletely understood. Here, using several human cell lines, immunofluorescence and structured illumination microscopy methods, and ubiquitination assays, we show that the E3 ubiquitin ligase F-box and WD repeat domain-containing 7 (FBXW7), a subunit of the SCF ubiquitin ligase, down-regulates spindle assembly 6 homolog (HsSAS-6), a key protein required for procentriole cartwheel assembly, and thereby regulates centriole duplication. We found that FBXW7 abrogation stabilizes HsSAS-6 and increases its recruitment to the mother centriole at multiple sites, leading to supernumerary centrioles. Ultrastructural analyses revealed that FBXW7 is broadly localized on the mother centriole and that its presence is reduced at the site where the HsSAS-6-containing procentriole is formed. This observation suggested that FBXW7 restricts procentriole assembly to a specific site to generate a single new centriole. In contrast, during HsSAS-6 overexpression, FBXW7 strongly associated with HsSAS-6 at the centriole. We also found that SCFFBXW7 interacts with HsSAS-6 and targets it for ubiquitin-mediated degradation. Further, we identified putative phosphodegron sites in HsSAS-6, whose substitutions rendered it insensitive to FBXW7-mediated degradation and control of centriole number. In summary, SCFFBXW7 targets HsSAS-6 for degradation and thereby controls centriole biogenesis by restraining HsSAS-6 recruitment to the mother centriole, a molecular mechanism that controls supernumerary centrioles/centrosomes and the maintenance of bipolar spindles. .
Keywords: E3 ubiquitin ligase; F-Box and WD repeat domain-containing 7 (FBXW7); cell division; centriole; centrosome; human SAS-6 centriolar assembly protein (HsSAS6); mitosis; mitotic spindle; procentriole; ubiquitin. V体育安卓版.
© 2020 Badarudeen et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures (V体育ios版)

"VSports手机版" References
-
- Levine M. S., Bakker B., Boeckx B., Moyett J., Lu J., Vitre B., Spierings D. C., Lansdorp P. M., Cleveland D. W., Lambrechts D., Foijer F., and Holland A. J. (2017) Centrosome amplification is sufficient to promote spontaneous tumorigenesis in mammals. Dev. Cell 40, 313–322.e5 10.1016/j.devcel.2016.12.022 - VSports在线直播 - DOI - PMC - PubMed
"VSports在线直播" Publication types
- "VSports在线直播" Actions
MeSH terms (V体育2025版)
- VSports在线直播 - Actions
- "V体育2025版" Actions
- Actions (V体育官网)
- VSports最新版本 - Actions
- "V体育安卓版" Actions
- "V体育ios版" Actions
- VSports注册入口 - Actions
- Actions (V体育官网)
Substances
- "V体育平台登录" Actions
- "VSports手机版" Actions
- V体育官网入口 - Actions
"V体育官网" LinkOut - more resources
Full Text Sources (VSports注册入口)

