V体育2025版 - Memory B Cell Activation, Broad Anti-influenza Antibodies, and Bystander Activation Revealed by Single-Cell Transcriptomics
- PMID: 31968262
- PMCID: PMC7891556
- DOI: 10.1016/j.celrep.2019.12.063
Memory B Cell Activation, Broad Anti-influenza Antibodies, and Bystander Activation Revealed by Single-Cell Transcriptomics
Abstract (VSports手机版)
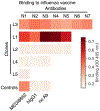
Antibody memory protects humans from many diseases. Protective antibody memory responses require activation of transcriptional programs, cell proliferation, and production of antigen-specific antibodies, but how these aspects of the response are coordinated is poorly understood VSports手机版. We profile the molecular and cellular features of the antibody response to influenza vaccination by integrating single-cell transcriptomics, longitudinal antibody repertoire sequencing, and antibody binding measurements. Single-cell transcriptional profiling reveals a program of memory B cell activation characterized by CD11c and T-bet expression associated with clonal expansion and differentiation toward effector function. Vaccination elicits an antibody clone, which rapidly acquired broad high-affinity hemagglutinin binding during affinity maturation. Unexpectedly, many antibody clones elicited by vaccination do not bind vaccine, demonstrating non-specific activation of bystander antibodies by influenza vaccination. These results offer insight into how molecular recognition, transcriptional programs, and clonal proliferation are coordinated in the human B cell repertoire during memory recall. .
Keywords: antibody repertoires; human antibody memory; infectious disease; influenza; single-cell transcriptomics; vaccination V体育安卓版. .
Copyright © 2019 V体育ios版. Published by Elsevier Inc. .
VSports注册入口 - Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures
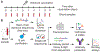



References
-
- Altschul SF, Gish W, Miller W, Myers EW, and Lipman DJ (1990). Basic local alignment search tool. J. Mol. Biol 215, 403–410. - PubMed
-
- Belshe RB, Newman FK, Cannon J, Duane C, Treanor J, Van Hoecke C, Howe BJ, and Dubin G (2004). Serum antibody responses after intradermal vaccination against influenza. N. Engl. J. Med 351, 2286–2294. - PubMed
-
- Bernasconi NL, Traggiai E, and Lanzavecchia A (2002). Maintenance of serological memory by polyclonal activation of human memory B cells. Science 298, 2199–2202. - PubMed
Publication types
- "V体育ios版" Actions
MeSH terms
- Actions (V体育安卓版)
- "VSports注册入口" Actions
- V体育ios版 - Actions
- "V体育官网入口" Actions
- "V体育ios版" Actions
- Actions (VSports最新版本)
- Actions (V体育官网)
- VSports在线直播 - Actions
- VSports在线直播 - Actions
Substances
- Actions (VSports app下载)
V体育官网入口 - Grants and funding
LinkOut - more resources
Full Text Sources
V体育官网入口 - Medical
Research Materials

