V体育官网 - An intra-tumoral niche maintains and differentiates stem-like CD8 T cells
- PMID: 31827286
- PMCID: PMC7108171
- DOI: VSports手机版 - 10.1038/s41586-019-1836-5
An intra-tumoral niche maintains and differentiates stem-like CD8 T cells
"VSports最新版本" Abstract
Tumour-infiltrating lymphocytes are associated with a survival benefit in several tumour types and with the response to immunotherapy1-8. However, the reason some tumours have high CD8 T cell infiltration while others do not remains unclear. Here we investigate the requirements for maintaining a CD8 T cell response against human cancer. We find that CD8 T cells within tumours consist of distinct populations of terminally differentiated and stem-like cells VSports手机版. On proliferation, stem-like CD8 T cells give rise to more terminally differentiated, effector-molecule-expressing daughter cells. For many T cells to infiltrate the tumour, it is critical that this effector differentiation process occur. In addition, we show that these stem-like T cells reside in dense antigen-presenting-cell niches within the tumour, and that tumours that fail to form these structures are not extensively infiltrated by T cells. Patients with progressive disease lack these immune niches, suggesting that niche breakdown may be a key mechanism of immune escape. .
Conflict of interest statement
"VSports最新版本" Figures






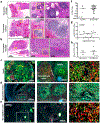

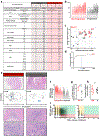




Comment in
-
Identifying the source of tumour-infiltrating T cells.Nature. 2019 Dec;576(7787):385-386. doi: 10.1038/d41586-019-03670-6. Nature. 2019. PMID: 31844254 No abstract available.
-
"V体育官网入口" The source within - intratumoural stem-like T cells give rise to differentiated T cells.Nat Rev Cancer. 2020 Mar;20(3):140. doi: 10.1038/s41568-020-0239-0. Nat Rev Cancer. 2020. PMID: 31925398 No abstract available.
V体育2025版 - References
-
- Galon J et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science 313, 1960–1964 (2006). - PubMed (VSports app下载)
-
- Pagès F et al. Effector memory T cells, early metastasis, and survival in colorectal cancer. N. Engl. J. Med 353, 2654–2666 (2005). - V体育ios版 - PubMed
-
- Azimi F et al. Tumor-infiltrating lymphocyte grade is an independent predictor of sentinel lymph node status and survival in patients with cutaneous melanoma. J. Clin. Oncol 30, 2678–2683 (2012). - PubMed
-
- Savas P et al. Single-cell profiling of breast cancer T cells reveals a tissue-resident memory subset associated with improved prognosis. Nat. Med 24, 986–993 (2018). - PubMed
Publication types
- Actions (V体育2025版)
"VSports app下载" MeSH terms
- Actions (V体育官网入口)
- "V体育官网" Actions
- VSports最新版本 - Actions
- VSports最新版本 - Actions
- "V体育官网" Actions
- VSports - Actions
Substances
- "V体育安卓版" Actions
- Actions (VSports app下载)
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

