Mechanical strain promotes skin fibrosis through LRG-1 induction mediated by ELK1 and ERK signalling
- PMID: 31602408
- PMCID: PMC6778114
- DOI: VSports注册入口 - 10.1038/s42003-019-0600-6
Mechanical strain promotes skin fibrosis through LRG-1 induction mediated by ELK1 and ERK signalling
"VSports注册入口" Abstract
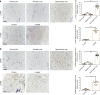
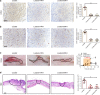
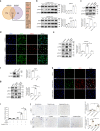
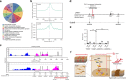
Biomechanical force and pathological angiogenesis are dominant features in fibro-proliferative disorders. Understanding the role and regulation of the mechanical microenvironment in which pathological angiogenesis occurs is an important challenge when investigating numerous angiogenesis-related diseases. In skin fibrosis, dermal fibroblasts and vascular endothelial cells are integral to hypertrophic scar formation. However, few studies have been conducted to closely investigate their relationship. Here we show, that leucine-rich-alpha-2-glycoprotein 1 (LRG-1) a regulator of pathological angiogenesis, links biomechanical force to angiogenesis in skin fibrosis. We discover that LRG-1 is overexpressed in hypertrophic scar tissues, and that depletion of Lrg-1 in mouse skin causes mild neovascularization and skin fibrosis formation in a hypertrophic scarring model. Inhibition of FAK or ERK attenuates LRG-1 expression through the ELK1 transcription factor, which binds to the LRG-1 promoter region after transcription initiation by mechanical force. Using LRG-1 to uncouple mechanical force from angiogenesis may prove clinically successful in treating fibro-proliferative disorders. VSports手机版.
Keywords: Angiogenesis; Cell signalling; Mechanisms of disease. V体育安卓版.
© The Author(s) 2019.
Conflict of interest statement
Competing interestsThe authors declare no competing interests.
Figures
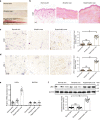
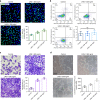
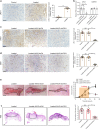

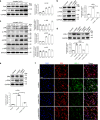
VSports手机版 - References
V体育ios版 - Publication types
MeSH terms
- Actions (V体育官网入口)
- VSports - Actions
- "V体育官网" Actions
- Actions (VSports手机版)
- "V体育安卓版" Actions
- V体育平台登录 - Actions
Substances
- "VSports手机版" Actions
- "V体育安卓版" Actions
- V体育ios版 - Actions
LinkOut - more resources
Full Text Sources
"V体育安卓版" Molecular Biology Databases
Miscellaneous

