"VSports在线直播" Metabolic enzyme PDK3 forms a positive feedback loop with transcription factor HSF1 to drive chemoresistance
- PMID: 31244938
- PMCID: PMC6568185
- DOI: 10.7150/thno.31301
Metabolic enzyme PDK3 forms a positive feedback loop with transcription factor HSF1 to drive chemoresistance (VSports app下载)
VSports注册入口 - Abstract
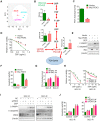
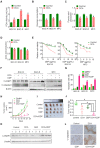
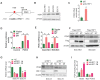
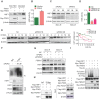
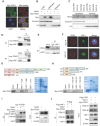
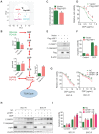
Background & Aims: Dysregulation of metabolism plays an important role in the development and progression of cancers, while the underlying mechanisms remain largely unknown. This study aims to explore the regulation and relevance of glycolysis in chemoresistance of gastric cancer. Methods: Biochemical differences between chemoresistant and chemosensitive cancer cells were determined by metabolism profiling, microarray gene expression, PCR or western blotting. Cancer cell growth in vitro or in vivo were analyzed by viability, apoptosis and nude mice assay. Immunoprecipation was used to explore the interaction of proteins with other proteins or DNAs. Results: By metabolic and gene expression profiling, we found that pyruvate dehydrogenase kinase 3 (PDK3) was highly expressed to promote glycolysis in chemoresistant cancer cells. Its genetic or chemical inhibition reverted chemoresistance in vitro and in vivo. It was transcriptionally regulated by transcription factor HSF1 (Heat shock factor 1). Interestingly, PDK3 can localize in the nucleus and interact with HSF1 to disrupt its phosphorylation by GSK3β VSports手机版. Since HSF1 was subjected to FBXW7-catalyzed polyubiquitination in a phosphorylation-dependent manner, PDK3 prevented HSF1 from proteasomal degradation. Thus, metabolic enzyme PDK3 and transcription factor HSF1 forms a positive feedback loop to promote glycolysis. As a result, inhibition of HSF1 impaired enhanced glycolysis and reverted chemoresistance both in vitro and in vivo. Conclusions: PDK3 forms a positive feedback loop with HSF1 to drive glycolysis in chemoresistance. Targeting this mitonuclear communication may represent a novel approach to overcome chemoresistance. .
Keywords: HSF1; PDK3; chemoresistance; glycolysis; metabolism. V体育安卓版.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures


References
-
- Ferreira JA, Peixoto A, Neves M, Gaiteiro C, Reis CA, Assaraf YG. et al. Mechanisms of cisplatin resistance and targeting of cancer stem cells: Adding glycosylation to the equation. Drug Resist Updat. 2016;24:34–54. - PubMed
-
- Liu X, Wang X, Zhang J, Lam EK, Shin VY, Cheng AS. et al. Warburg effect revisited: an epigenetic link between glycolysis and gastric carcinogenesis. Oncogene. 2010;29:442–50. - PubMed
"V体育官网" Publication types
MeSH terms
- VSports注册入口 - Actions
- V体育2025版 - Actions
- "V体育ios版" Actions
- VSports注册入口 - Actions
- "V体育ios版" Actions
- "VSports注册入口" Actions
- V体育2025版 - Actions
- "VSports最新版本" Actions
- Actions (VSports在线直播)
Substances (VSports在线直播)
- Actions (V体育2025版)
- Actions (VSports注册入口)
- Actions (VSports最新版本)
- V体育2025版 - Actions
VSports app下载 - LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous (V体育官网)

