The IRE1 endoplasmic reticulum stress sensor activates natural killer cell immunity in part by regulating c-Myc
- PMID: 31086333
- PMCID: PMC6588410
- DOI: 10.1038/s41590-019-0388-z
The IRE1 endoplasmic reticulum stress sensor activates natural killer cell immunity in part by regulating c-Myc
"V体育ios版" Abstract
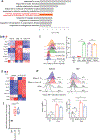
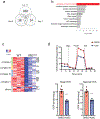
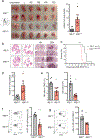
Natural killer (NK) cells are critical mediators of host immunity to pathogens. Here, we demonstrate that the endoplasmic reticulum stress sensor inositol-requiring enzyme 1 (IRE1α) and its substrate transcription factor X-box-binding protein 1 (XBP1) drive NK cell responses against viral infection and tumors in vivo. IRE1α-XBP1 were essential for expansion of activated mouse and human NK cells and are situated downstream of the mammalian target of rapamycin signaling pathway. Transcriptome and chromatin immunoprecipitation analysis revealed c-Myc as a new and direct downstream target of XBP1 for regulation of NK cell proliferation. Genetic ablation or pharmaceutical blockade of IRE1α downregulated c-Myc, and NK cells with c-Myc haploinsufficency phenocopied IRE1α-XBP1 deficiency. c-Myc overexpression largely rescued the proliferation defect in IRE1α-/- NK cells. Like c-Myc, IRE1α-XBP1 also promotes oxidative phosphorylation in NK cells. Overall, our study identifies a IRE1α-XBP1-cMyc axis in NK cell immunity, providing insight into host protection against infection and cancer VSports手机版. .
Conflict of interest statement
Competing interests
L. H. G V体育安卓版. is a former Director of Bristol-Myers Squibb and is currently on the board of directors of and holds equity in GlaxoSmithKline Pharmaceuticals and the Waters Corporation. She chairs the scientific advisory board, is a co-founder of and holds equity in Quentis Therapeutics. She also serves on the scientific advisory boards of Repare Therapeutics, Abro and Kaleido Therapeutics.
Figures





References
References (for main text only)
-
- Sun JC et al. Proinflammatory cytokine signaling required for the generation of natural killer cell memory. J Exp Med 209, 947–954 (2012). - VSports app下载 - PMC - PubMed
References (Methods-only)
-
- Anders S & Huber W Differential expression analysis for sequence count data. Genome Biol 11, R106 (2010). - V体育2025版 - PMC - PubMed
Publication types
MeSH terms
- Actions (V体育2025版)
- "V体育平台登录" Actions
- V体育官网入口 - Actions
- VSports注册入口 - Actions
- VSports注册入口 - Actions
- "V体育2025版" Actions
- Actions (V体育官网入口)
- Actions (VSports注册入口)
- Actions (V体育2025版)
- Actions (VSports在线直播)
- V体育平台登录 - Actions
- "V体育平台登录" Actions
Substances
- V体育官网入口 - Actions
- VSports注册入口 - Actions
V体育平台登录 - Grants and funding
LinkOut - more resources
"VSports手机版" Full Text Sources
"V体育安卓版" Other Literature Sources
"V体育2025版" Molecular Biology Databases
Research Materials

