Serum biomarkers identification by iTRAQ and verification by MRM: S100A8/S100A9 levels predict tumor-stroma involvement and prognosis in Glioblastoma
- PMID: 30808902
- PMCID: PMC6391445
- DOI: 10.1038/s41598-019-39067-8
"VSports app下载" Serum biomarkers identification by iTRAQ and verification by MRM: S100A8/S100A9 levels predict tumor-stroma involvement and prognosis in Glioblastoma
Abstract
Despite advances in biology and treatment modalities, the prognosis of glioblastoma (GBM) remains poor. Serum reflects disease macroenvironment and thus provides a less invasive means to diagnose and monitor a diseased condition. By employing 4-plex iTRAQ methodology, we identified 40 proteins with differential abundance in GBM sera. The high abundance of serum S100A8/S100A9 was verified by multiple reaction monitoring (MRM). ELISA and MRM-based quantitation showed a significant positive correlation. Further, an integrated investigation using stromal, tumor purity and cell type scores demonstrated an enrichment of myeloid cell lineage in the GBM tumor microenvironment. Transcript levels of S100A8/S100A9 were found to be independent poor prognostic indicators in GBM. Medium levels of pre-operative and three-month post-operative follow-up serum S100A8 levels predicted poor prognosis in GBM patients who lived beyond median survival. In vitro experiments showed that recombinant S100A8/S100A9 proteins promoted integrin signalling dependent glioma cell migration and invasion up to a threshold level of concentrations. Thus, we have discovered GBM serum marker by iTRAQ and verified by MRM. We also demonstrate interplay between tumor micro and macroenvironment and identified S100A8 as a potential marker with diagnostic and prognostic value in GBM. VSports手机版.
Conflict of interest statement
The authors declare no competing interests.
Figures
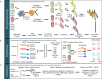



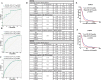
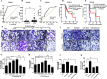
References
-
- Stupp R, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. The Lancet. Oncology. 2009;10:459–466. doi: 10.1016/S1470-2045(09)70025-7. - DOI - PubMed
-
- Tanwar MK, Gilbert MR, Holland EC. Gene expression microarray analysis reveals YKL-40 to be a potential serum marker for malignant character in human glioma. Cancer research. 2002;62:4364–4368. - V体育安卓版 - PubMed
-
- Jung CS, et al. Serum GFAP is a diagnostic marker for glioblastoma multiforme. Brain: a journal of neurology. 2007;130:3336–3341. doi: 10.1093/brain/awm263. - DOI (VSports注册入口) - PubMed
V体育ios版 - Publication types
- "VSports在线直播" Actions
MeSH terms (VSports)
- "V体育平台登录" Actions
- "V体育2025版" Actions
- "V体育2025版" Actions
- VSports在线直播 - Actions
- "VSports" Actions
- "V体育平台登录" Actions
- VSports手机版 - Actions
- Actions (VSports注册入口)
"VSports在线直播" Substances
- V体育平台登录 - Actions
- Actions (V体育2025版)
- "V体育官网入口" Actions
LinkOut - more resources
Full Text Sources
Miscellaneous

