LUBAC determines chemotherapy resistance in squamous cell lung cancer (VSports手机版)
- PMID: 30642944
- PMCID: PMC6363428
- DOI: "VSports手机版" 10.1084/jem.20180742
LUBAC determines chemotherapy resistance in squamous cell lung cancer
V体育官网 - Abstract
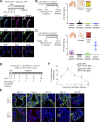
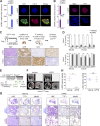
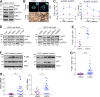
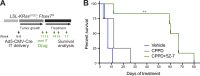
Lung squamous cell carcinoma (LSCC) and adenocarcinoma (LADC) are the most common lung cancer subtypes. Molecular targeted treatments have improved LADC patient survival but are largely ineffective in LSCC. The tumor suppressor FBW7 is commonly mutated or down-regulated in human LSCC, and oncogenic KRasG12D activation combined with Fbxw7 inactivation in mice (KF model) caused both LSCC and LADC. Lineage-tracing experiments showed that CC10+, but not basal, cells are the cells of origin of LSCC in KF mice VSports手机版. KF LSCC tumors recapitulated human LSCC resistance to cisplatin-based chemotherapy, and we identified LUBAC-mediated NF-κB signaling as a determinant of chemotherapy resistance in human and mouse. Inhibition of NF-κB activation using TAK1 or LUBAC inhibitors resensitized LSCC tumors to cisplatin, suggesting a future avenue for LSCC patient treatment. .
© 2019 Ruiz et al.
Figures (VSports注册入口)




References
-
- Campbell J.D., Alexandrov A., Kim J., Wala J., Berger A.H., Pedamallu C.S., Shukla S.A., Guo G., Brooks A.N., Murray B.A., et al. Cancer Genome Atlas Research Network . 2016. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat. Genet. 48:607–616. 10.1038/ng.3564 - "VSports手机版" DOI - PMC - PubMed
-
- Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., Jacobsen A., Byrne C.J., Heuer M.L., Larsson E., et al. . 2012. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2:401–404. 10.1158/2159-8290.CD-12-0095 - DOI - PMC - PubMed
Publication types
MeSH terms
- VSports在线直播 - Actions
- "V体育2025版" Actions
- Actions (VSports注册入口)
- Actions (V体育平台登录)
- "VSports app下载" Actions
- V体育官网入口 - Actions
- V体育2025版 - Actions
- Actions (VSports)
Substances
- "V体育官网" Actions
- VSports手机版 - Actions
- "VSports手机版" Actions
Grants and funding
"VSports手机版" LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

