Enriching Beneficial Microbial Diversity of Indoor Plants and Their Surrounding Built Environment With Biostimulants
- PMID: 30568641
- PMCID: PMC6290261
- DOI: 10.3389/fmicb.2018.02985
Enriching Beneficial Microbial Diversity of Indoor Plants and Their Surrounding Built Environment With Biostimulants
VSports最新版本 - Abstract
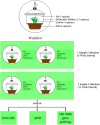
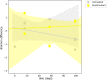
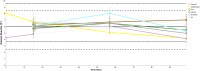
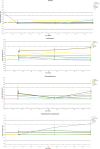
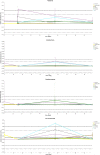
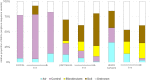
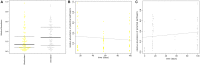
Microbial diversity is suggested as the key for plant and human health VSports手机版. However, how microbial diversity can be enriched is largely unknown but of great interest for health issues. Biostimulants offer the way to directly augment our main living areas by the healthy microbiome of indoor plants. Here, we investigated shifts of the microbiome on leaves of spider plants (Chlorophytum comosum) and its surrounding abiotic surfaces in the built environment after irrigation with a vermicompost-based biostimulant for 12 weeks. The biostimulant could not only promote plant growth, but changed the composition of the microbiome and abundance of intact microbial cells on plant leaves and even stronger on abiotic surfaces in close vicinity under constant conditions of the microclimate. Biostimulant treatments stabilized microbial diversity and resulted in an increase of Bacteroidetes and a surprising transient emerge of new phyla, e. g. , Verrucomicrobia, Acidobacteria, and Thaumarchaeota. The proportion of potentially beneficial microorganisms like Brevibacillus, Actinoallomurus, Paenibacillus, Sphaerisporangium increased relatively; microbial diversity was stabilized, and the built environment became more plant-like. Detected metabolites like indole-3-acetic acid in the biostimulant were potentially contributed by species of Pseudomonas. Overall, effects of the biostimulant on the composition of the microbiome could be predicted with an accuracy of 87%. This study shows the potential of biostimulants not only for the plant itself, but also for other living holobionts like humans in the surrounding environment. .
Keywords: 16S rRNA gene amplicon analysis; LC-MS; biostimulants; built environment; indoor plants; microbiome; qPCR; vermicompost. V体育安卓版.
Figures


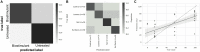


"VSports" References
-
- Anderson M. J. (2001). A new method for non-parametric multivariate analysis of variance. Austral Ecol. 26 32–46.
-
- Berg G. (2009). Plant-microbe interactions promoting plant growth and health: perspectives for controlled use of microorganisms in agriculture. Appl. Microbiol. Biotechnol. 84 11–18. 10.1007/s00253-009-2092-7 - "V体育官网" DOI - PubMed
Grants and funding
LinkOut - more resources
VSports在线直播 - Full Text Sources

