Metabolic heterogeneity underlies reciprocal fates of TH17 cell stemness and plasticity
- PMID: 30568299
- PMCID: PMC6420879
- DOI: 10.1038/s41586-018-0806-7
Metabolic heterogeneity underlies reciprocal fates of TH17 cell stemness and plasticity
Abstract
A defining feature of adaptive immunity is the development of long-lived memory T cells to curtail infection. Recent studies have identified a unique stem-like T-cell subset amongst exhausted CD8-positive T cells in chronic infection1-3, but it remains unclear whether CD4-positive T-cell subsets with similar features exist in chronic inflammatory conditions VSports手机版. Amongst helper T cells, TH17 cells have prominent roles in autoimmunity and tissue inflammation and are characterized by inherent plasticity4-7, although how such plasticity is regulated is poorly understood. Here we demonstrate that TH17 cells in a mouse model of autoimmune disease are functionally and metabolically heterogeneous; they contain a subset with stemness-associated features but lower anabolic metabolism, and a reciprocal subset with higher metabolic activity that supports transdifferentiation into TH1-like cells. These two TH17-cell subsets are defined by selective expression of the transcription factors TCF-1 and T-bet, and by discrete levels of CD27 expression. We also identify signalling via the kinase complex mTORC1 as a central regulator of TH17-cell fate decisions by coordinating metabolic and transcriptional programmes. TH17 cells with disrupted mTORC1 signalling or anabolic metabolism fail to induce autoimmune neuroinflammation or to develop into TH1-like cells, but instead upregulate TCF-1 expression and acquire stemness-associated features. Single-cell RNA sequencing and experimental validation reveal heterogeneity in fate-mapped TH17 cells, and a developmental arrest in the TH1 transdifferentiation trajectory upon loss of mTORC1 activity or metabolic perturbation. Our results establish that the dichotomy of stemness and effector function underlies the heterogeneous TH17 responses and autoimmune pathogenesis, and point to previously unappreciated metabolic control of plasticity in helper T cells. .
"VSports最新版本" Figures










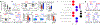
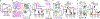

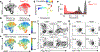
Comment in (V体育官网入口)
-
mTOR Is Key to T Cell Transdifferentiation.Cell Metab. 2019 Feb 5;29(2):241-242. doi: 10.1016/j.cmet.2019.01.008. Cell Metab. 2019. PMID: 30726756 Free PMC article.
"VSports" References
-
- Leong YA et al. CXCR5(+) follicular cytotoxic T cells control viral infection in B cell follicles. Nat. Immunol. 17, 1187–1196, doi:10.1038/ni.3543 (2016). - DOI (VSports app下载) - PubMed
MeSH terms
- "V体育ios版" Actions
- "V体育官网" Actions
- Actions (V体育2025版)
- Actions (VSports注册入口)
- "VSports在线直播" Actions
- V体育安卓版 - Actions
- Actions (V体育安卓版)
- "VSports在线直播" Actions
- V体育安卓版 - Actions
- "V体育平台登录" Actions
- Actions (V体育官网)
- Actions (V体育官网入口)
VSports - Substances
- "V体育安卓版" Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
V体育ios版 - Medical
Molecular Biology Databases
Research Materials

