KDM4B-regulated unfolded protein response as a therapeutic vulnerability in PTEN-deficient breast cancer (V体育平台登录)
- PMID: 30266800
- PMCID: "VSports手机版" PMC6219741
- DOI: 10.1084/jem.20180439
KDM4B-regulated unfolded protein response as a therapeutic vulnerability in PTEN-deficient breast cancer (VSports在线直播)
Abstract (V体育ios版)
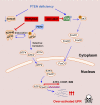
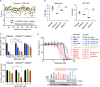
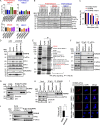
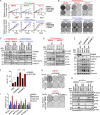
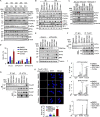
PTEN deficiency in breast cancer leads to resistance to PI3K-AKT inhibitor treatment despite aberrant activation of this signaling pathway. Here, we report that genetic depletion or small molecule inhibition of KDM4B histone demethylase activates the unfolded protein response (UPR) pathway and results in preferential apoptosis in PTEN-deficient triple-negative breast cancers (TNBCs). Intriguingly, this function of KDM4B on UPR requires its demethylase activity but is independent of its canonical role in histone modification, and acts through its cytoplasmic interaction with eIF2α, a crucial component of UPR signaling, resulting in reduced phosphorylation of this component VSports手机版. Targeting KDM4B in combination with PI3K inhibition induces further activation of UPR, leading to robust synergy in apoptosis. These findings identify KDM4B as a therapeutic vulnerability in PTEN-deficient TNBC that otherwise would be resistant to PI3K inhibition. .
© 2018 Wang et al.
"V体育官网入口" Figures



References
-
- Benitez J.A., Ma J., D’Antonio M., Boyer A., Camargo M.F., Zanca C., Kelly S., Khodadadi-Jamayran A., Jameson N.M., Andersen M., et al. . 2017. PTEN regulates glioblastoma oncogenesis through chromatin-associated complexes of DAXX and histone H3.3. Nat. Commun. 8:15223 10.1038/ncomms15223 - DOI - PMC - PubMed
-
- Berry W.L., Kim T.D., and Janknecht R.. 2014. Stimulation of β-catenin and colon cancer cell growth by the KDM4B histone demethylase. Int. J. Oncol. 44:1341–1348. 10.3892/ijo.2014.2279 - V体育官网 - DOI - PubMed
-
- Beyer S., Kristensen M.M., Jensen K.S., Johansen J.V., and Staller P.. 2008. The histone demethylases JMJD1A and JMJD2B are transcriptional targets of hypoxia-inducible factor HIF. J. Biol. Chem. 283:36542–36552. 10.1074/jbc.M804578200 - DOI (VSports) - PMC - PubMed
-
- Brachmann S.M., Hofmann I., Schnell C., Fritsch C., Wee S., Lane H., Wang S., Garcia-Echeverria C., and Maira S.M.. 2009. Specific apoptosis induction by the dual PI3K/mTor inhibitor NVP-BEZ235 in HER2 amplified and PIK3CA mutant breast cancer cells. Proc. Natl. Acad. Sci. USA. 106:22299–22304. 10.1073/pnas.0905152106 - "VSports在线直播" DOI - PMC - PubMed
-
- Cancer Genome Atlas Network 2012. Comprehensive molecular portraits of human breast tumours. Nature. 490:61–70. 10.1038/nature11412 - "VSports手机版" DOI - PMC - PubMed
Publication types
- "VSports在线直播" Actions
MeSH terms
- "VSports最新版本" Actions
- "VSports在线直播" Actions
- Actions (VSports app下载)
- Actions (VSports手机版)
- "VSports app下载" Actions
- "VSports" Actions
- Actions (VSports)
- Actions (V体育官网)
- V体育ios版 - Actions
- Actions (VSports注册入口)
- V体育官网入口 - Actions
- Actions (VSports手机版)
- "VSports app下载" Actions
- V体育官网入口 - Actions
- Actions (V体育ios版)
- "V体育平台登录" Actions
- "V体育平台登录" Actions
- "VSports" Actions
Substances
- V体育平台登录 - Actions
- "V体育2025版" Actions
- Actions (V体育ios版)
- V体育平台登录 - Actions
- VSports注册入口 - Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

