Differential IL-2 expression defines developmental fates of follicular versus nonfollicular helper T cells (V体育2025版)
- PMID: 30213884
- PMCID: PMC6501592
- DOI: 10.1126/science.aao2933
Differential IL-2 expression defines developmental fates of follicular versus nonfollicular helper T cells (VSports手机版)
"VSports在线直播" Abstract
In response to infection, naïve CD4+ T cells differentiate into two subpopulations: T follicular helper (TFH) cells, which support B cell antibody production, and non-TFH cells, which enhance innate immune cell functions. Interleukin-2 (IL-2), the major cytokine produced by naïve T cells, plays an important role in the developmental divergence of these populations. However, the relationship between IL-2 production and fate determination remains unclear. Using reporter mice, we found that differential production of IL-2 by naïve CD4+ T cells defined precursors fated for different immune functions. IL-2 producers, which were fated to become TFH cells, delivered IL-2 to nonproducers destined to become non-TFH cells VSports手机版. Because IL-2 production was limited to cells receiving the strongest T cell receptor (TCR) signals, a direct link between TCR-signal strength, IL-2 production, and T cell fate determination has been established. .
Copyright © 2018 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U. S V体育安卓版. Government Works. .
"VSports在线直播" Figures

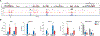





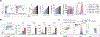
References
-
- Bucy RP et al., Single cell analysis of cytokine gene coexpression during CD4+ T-cell phenotype development. Proc. Nat.l Acad. Sci. USA 92, 7565–7569 (1995). - "V体育官网入口" PMC - PubMed
-
- Nurieva RI et al., STAT5 protein negatively regulates T follicular helper (Tfh) cell generation and function. J Biol. Chem 287, 11234–11239 (2012). - VSports手机版 - PMC - PubMed
Publication types
- "VSports手机版" Actions
MeSH terms
- "VSports最新版本" Actions
- V体育官网 - Actions
- "VSports" Actions
- "V体育ios版" Actions
- Actions (V体育安卓版)
VSports app下载 - Substances
- "VSports" Actions
- VSports在线直播 - Actions
"VSports最新版本" Grants and funding
- R01 AI120202/AI/NIAID NIH HHS/United States
- T32 GM008169/GM/NIGMS NIH HHS/United States
- P30 DK043351/DK/NIDDK NIH HHS/United States
- P30 CA013148/CA/NCI NIH HHS/United States
- R01 AI124657/AI/NIAID NIH HHS/United States
- R21 AI124143/AI/NIAID NIH HHS/United States
- R01 AI107020/AI/NIAID NIH HHS/United States (VSports)
- R01 AI110113/AI/NIAID NIH HHS/United States
- V体育安卓版 - F30 DK105680/DK/NIDDK NIH HHS/United States
- R21 AI035783/AI/NIAID NIH HHS/United States
- R01 DK113789/DK/NIDDK NIH HHS/United States
- R01 AI035783/AI/NIAID NIH HHS/United States
- T32 AI007051/AI/NIAID NIH HHS/United States
- R01 AI057956/AI/NIAID NIH HHS/United States (V体育ios版)
- VSports手机版 - R01 AI077574/AI/NIAID NIH HHS/United States
- DP1 AI131080/AI/NIAID NIH HHS/United States
- VSports app下载 - T32 GM008361/GM/NIGMS NIH HHS/United States
V体育安卓版 - LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases (V体育官网入口)
"V体育官网" Research Materials
V体育ios版 - Miscellaneous

