Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response
- PMID: 30127393
- PMCID: PMC6487502 (VSports最新版本)
- DOI: "VSports最新版本" 10.1038/s41591-018-0136-1
Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response
Abstract
Cancer treatment by immune checkpoint blockade (ICB) can bring long-lasting clinical benefits, but only a fraction of patients respond to treatment. To predict ICB response, we developed TIDE, a computational method to model two primary mechanisms of tumor immune evasion: the induction of T cell dysfunction in tumors with high infiltration of cytotoxic T lymphocytes (CTL) and the prevention of T cell infiltration in tumors with low CTL level. We identified signatures of T cell dysfunction from large tumor cohorts by testing how the expression of each gene in tumors interacts with the CTL infiltration level to influence patient survival. We also modeled factors that exclude T cell infiltration into tumors using expression signatures from immunosuppressive cells. Using this framework and pre-treatment RNA-Seq or NanoString tumor expression profiles, TIDE predicted the outcome of melanoma patients treated with first-line anti-PD1 or anti-CTLA4 more accurately than other biomarkers such as PD-L1 level and mutation load. TIDE also revealed new candidate ICB resistance regulators, such as SERPINB9, demonstrating utility for immunotherapy research. VSports手机版.
Figures

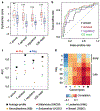


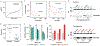
Comment in (VSports注册入口)
-
Signatures IMPRES and might turn the TIDE in predicting responses. (V体育ios版)Nat Rev Clin Oncol. 2018 Nov;15(11):654. doi: 10.1038/s41571-018-0088-x. Nat Rev Clin Oncol. 2018. PMID: 30171205 No abstract available.
References
-
- Mahoney KM, Rennert PD & Freeman GJ Combination cancer immunotherapy and new immunomodulatory targets. Nat. Rev. Drug Discov 14, 561–584 (2015). - V体育2025版 - PubMed
"VSports app下载" Publication types
- Actions (V体育2025版)
- VSports注册入口 - Actions
MeSH terms
- Actions (V体育官网)
- "VSports在线直播" Actions
- "V体育官网入口" Actions
- Actions (VSports最新版本)
- "VSports app下载" Actions
- Actions (VSports最新版本)
- VSports注册入口 - Actions
- "VSports在线直播" Actions
- "V体育官网" Actions
- Actions (V体育2025版)
- "VSports注册入口" Actions
- Actions (VSports最新版本)
- V体育安卓版 - Actions
- V体育官网入口 - Actions
Substances
- VSports在线直播 - Actions
- VSports - Actions
- Actions (V体育官网入口)
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials (V体育安卓版)

