Functional Proteomic Profiling of Secreted Serine Proteases in Health and Inflammatory Bowel Disease
- PMID: 29777136
- PMCID: PMC5959920
- DOI: 10.1038/s41598-018-26282-y
Functional Proteomic Profiling of Secreted Serine Proteases in Health and Inflammatory Bowel Disease
Abstract
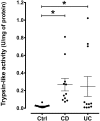
While proteases are essential in gastrointestinal physiology, accumulating evidence indicates that dysregulated proteolysis plays a pivotal role in the pathophysiology of inflammatory bowel disease (IBD) VSports手机版. Nonetheless, the identity of overactive proteases released by human colonic mucosa remains largely unknown. Studies of protease abundance have primarily investigated expression profiles, not taking into account their enzymatic activity. Herein we have used serine protease-targeted activity-based probes (ABPs) coupled with mass spectral analysis to identify active forms of proteases secreted by the colonic mucosa of healthy controls and IBD patients. Profiling of (Pro-Lys)-ABP bound proteases revealed that most of hyperactive proteases from IBD secretome are clustered at 28-kDa. We identified seven active proteases: the serine proteases cathepsin G, plasma kallikrein, plasmin, tryptase, chymotrypsin-like elastase 3 A, and thrombin and the aminopeptidase B. Only cathepsin G and thrombin were overactive in supernatants from IBD patient tissues compared to healthy controls. Gene expression analysis highlighted the transcription of genes encoding these proteases into intestinal mucosae. The functional ABP-targeted proteomic approach that we have used to identify active proteases in human colonic samples bears directly on the understanding of the role these enzymes may play in the pathophysiology of IBD. .
Conflict of interest statement
The authors declare no competing interests.
Figures



References
"V体育官网入口" Publication types
MeSH terms (V体育官网)
- Actions (VSports最新版本)
- VSports app下载 - Actions
- "V体育官网入口" Actions
- Actions (VSports最新版本)
- Actions (VSports app下载)
- Actions (V体育2025版)
- "VSports手机版" Actions
- "VSports手机版" Actions
- "VSports" Actions
- VSports app下载 - Actions
Substances
VSports最新版本 - LinkOut - more resources
"V体育官网" Full Text Sources
V体育ios版 - Other Literature Sources
Medical
VSports最新版本 - Molecular Biology Databases

