Distinct chromatin functional states correlate with HIV latency reactivation in infected primary CD4+ T cells
- PMID: 29714165
- PMCID: PMC5973828
- DOI: 10.7554/eLife.34655 (V体育ios版)
Distinct chromatin functional states correlate with HIV latency reactivation in infected primary CD4+ T cells
Abstract
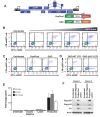
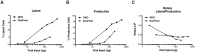
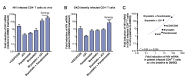
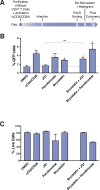
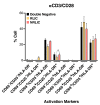
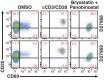
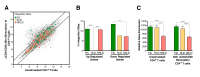
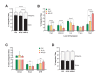
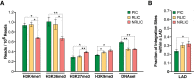
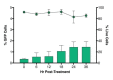
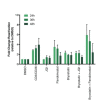
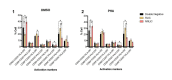
Human immunodeficiency virus (HIV) infection is currently incurable, due to the persistence of latently infected cells. The 'shock and kill' approach to a cure proposes to eliminate this reservoir via transcriptional activation of latent proviruses, enabling direct or indirect killing of infected cells. Currently available latency-reversing agents (LRAs) have however proven ineffective. To understand why, we used a novel HIV reporter strain in primary CD4+ T cells and determined which latently infected cells are reactivatable by current candidate LRAs. Remarkably, none of these agents reactivated more than 5% of cells carrying a latent provirus VSports手机版. Sequencing analysis of reactivatable vs. non-reactivatable populations revealed that the integration sites were distinguishable in terms of chromatin functional states. Our findings challenge the feasibility of 'shock and kill', and suggest the need to explore other strategies to control the latent HIV reservoir. .
Keywords: HIV-1 latency; human; infectious disease; integration sites; latency reversal; latency reversing agents; microbiology; reservoirs V体育安卓版. .
© 2018, Battivelli et al.
Conflict of interest statement (V体育安卓版)
EB, MD, MA, JS, IT, LC, AG, SD, WG, SP, EV No competing interests declared
Figures

Comment in
-
"V体育官网入口" Coloring hidden viruses.Elife. 2018 May 29;7:e37732. doi: 10.7554/eLife.37732. Elife. 2018. PMID: 29809152 Free PMC article.
"V体育平台登录" References
-
- Archin NM, Liberty AL, Kashuba AD, Choudhary SK, Kuruc JD, Crooks AM, Parker DC, Anderson EM, Kearney MF, Strain MC, Richman DD, Hudgens MG, Bosch RJ, Coffin JM, Eron JJ, Hazuda DJ, Margolis DM. Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature. 2012a;487:482–485. doi: 10.1038/nature11286. - DOI - PMC - PubMed
-
- Archin NM, Vaidya NK, Kuruc JD, Liberty AL, Wiegand A, Kearney MF, Cohen MS, Coffin JM, Bosch RJ, Gay CL, Eron JJ, Margolis DM, Perelson AS. Immediate antiviral therapy appears to restrict resting CD4+ cell HIV-1 infection without accelerating the decay of latent infection. PNAS. 2012b;109:9523–9528. doi: 10.1073/pnas.1120248109. - VSports注册入口 - DOI - PMC - PubMed
-
- Battivelli E, Verdin E. HIVGKO: a tool to assess HIV-1 latency reversal agents in human primary CD4+ T cells. Bio-Protocol. 2018;8:e3050. doi: 10.21769/BioProtoc.3050. - DOI (V体育安卓版) - PMC - PubMed
Publication types
- "VSports" Actions
- VSports app下载 - Actions
MeSH terms
- Actions (VSports最新版本)
- "V体育平台登录" Actions
- "VSports手机版" Actions
- V体育官网 - Actions
- Actions (V体育ios版)
- Actions (VSports注册入口)
VSports最新版本 - Substances
Grants and funding
- VSports最新版本 - T32 AI070084/AI/NIAID NIH HHS/United States
- 201311MFE-321128-179658/CIHR/Canada
- R01GM117901/GM/NIGMS NIH HHS/United States
- "VSports" DP1 DA031126/DA/NIDA NIH HHS/United States
- R01 GM117901/GM/NIGMS NIH HHS/United States
- S10 RR028962/RR/NCRR NIH HHS/United States
- R21 AI129636/AI/NIAID NIH HHS/United States
- R21AI129636/AI/NIAID NIH HHS/United States
- R01 DA041742/DA/NIDA NIH HHS/United States
- 1R01DE026010-01/DE/NIDCR NIH HHS/United States
- R21 AI143385/AI/NIAID NIH HHS/United States
- P30 CA010815/CA/NCI NIH HHS/United States
- 5-31532/DE/NIDCR NIH HHS/United States
- R01 AI150449/AI/NIAID NIH HHS/United States
- P30 AI027763/AI/NIAID NIH HHS/United States
- R01Ai117864/AI/NIAID NIH HHS/United States
- R01 DE026010/DE/NIDCR NIH HHS/United States
- R01 DA030216/DA/NIDA NIH HHS/United States
- "VSports注册入口" 1R01DA041742-01/DA/NIDA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials (V体育2025版)
