Loss of KLHL6 promotes diffuse large B-cell lymphoma growth and survival by stabilizing the mRNA decay factor roquin2 (VSports在线直播)
- PMID: 29695787
- PMCID: PMC5926793
- DOI: "VSports手机版" 10.1038/s41556-018-0084-5
Loss of KLHL6 promotes diffuse large B-cell lymphoma growth and survival by stabilizing the mRNA decay factor roquin2
Abstract
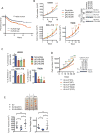
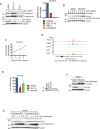
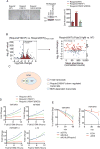
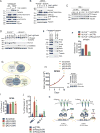
Kelch-like protein 6 (KLHL6) is an uncharacterized gene mutated in diffuse large B-cell lymphoma (DLBCL). Here we report that KLHL6 assembles with cullin3 to form a functional cullin-RING ubiquitin ligase. Mutations in KLHL6 inhibit its ligase activity by disrupting the interaction with cullin3. Loss of KLHL6 favours DLBCL growth and survival both in vitro and in xenograft models VSports手机版. We further established that the mRNA decay factor roquin2 is a substrate of KLHL6. Degradation of roquin2 is dependent on B-cell receptor activation, and requires the integrity of the Tyr691 residue in roquin2 that is essential for its interaction with KLHL6. A non-degradable roquin2(Y691F) mutant requires its RNA-binding ability to phenocopy the effect of KLHL6 loss. Stabilization of roquin2 promotes mRNA decay of the tumour suppressor and NF-κB pathway inhibitor, tumour necrosis factor-α-inducible gene 3. Collectively, our findings uncover the tumour suppressing mechanism of KLHL6. .
Conflict of interest statement
Author Information. The authors declare no competing financial interests V体育安卓版.
Figures (VSports手机版)



"V体育官网" References
-
- Gupta-Rossi N, et al. Specific over-expression of deltex and a new Kelch-like protein in human germinal center B cells. Mol Immunol. 2003;39:791–799. - PubMed
-
- Lohr JG, et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc Natl Acad Sci U S A. 2012;109:3879–3884. - "VSports最新版本" PMC - PubMed
Publication types
- "VSports app下载" Actions
MeSH terms
- Actions (VSports app下载)
- V体育ios版 - Actions
- Actions (V体育2025版)
- "V体育2025版" Actions
- V体育安卓版 - Actions
- Actions (VSports)
- "V体育ios版" Actions
- Actions (V体育ios版)
- Actions (V体育ios版)
- "V体育平台登录" Actions
- V体育安卓版 - Actions
Substances
- Actions (V体育2025版)
- "V体育ios版" Actions
- V体育安卓版 - Actions
- VSports最新版本 - Actions
- "V体育官网入口" Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

