Multi-stage Differentiation Defines Melanoma Subtypes with Differential Vulnerability to Drug-Induced Iron-Dependent Oxidative Stress (VSports app下载)
- PMID: 29657129
- PMCID: PMC5953834 (VSports)
- DOI: 10.1016/j.ccell.2018.03.017
"VSports app下载" Multi-stage Differentiation Defines Melanoma Subtypes with Differential Vulnerability to Drug-Induced Iron-Dependent Oxidative Stress
Abstract
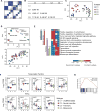
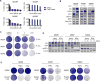
Malignant transformation can result in melanoma cells that resemble different stages of their embryonic development. Our gene expression analysis of human melanoma cell lines and patient tumors revealed that melanoma follows a two-dimensional differentiation trajectory that can be subclassified into four progressive subtypes. This differentiation model is associated with subtype-specific sensitivity to iron-dependent oxidative stress and cell death known as ferroptosis VSports手机版. Receptor tyrosine kinase-mediated resistance to mitogen-activated protein kinase targeted therapies and activation of the inflammatory signaling associated with immune therapy involves transitions along this differentiation trajectory, which lead to increased sensitivity to ferroptosis. Therefore, ferroptosis-inducing drugs present an orthogonal therapeutic approach to target the differentiation plasticity of melanoma cells to increase the efficacy of targeted and immune therapies. .
Keywords: combination therapy; differentiation; ferroptosis; immunotherapy; kinase inhibitor therapy; melanoma; pharmacogenomics; systems biology V体育安卓版. .
Copyright © 2018 Elsevier Inc V体育ios版. All rights reserved. .
VSports手机版 - Figures

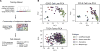



Comment in
-
Targeting the (Un)differentiated State of Cancer.Cancer Cell. 2018 May 14;33(5):793-795. doi: 10.1016/j.ccell.2018.04.007. Cancer Cell. 2018. PMID: 29763619
-
Four! Drivers of melanoma differentiation-When to use iron.Pigment Cell Melanoma Res. 2018 Nov;31(6):658-660. doi: 10.1111/pcmr.12725. Epub 2018 Aug 2. Pigment Cell Melanoma Res. 2018. PMID: 30015373 No abstract available.
"V体育官网入口" References
-
- Caramel J, Papadogeorgakis E, Hill L, Browne GJ, Richard G, Wierinckx A, Saldanha G, Osborne J, Hutchinson P, Tse G, et al. A Switch in the Expression of Embryonic EMT-Inducers Drives the Development of Malignant Melanoma. Cancer Cell. 2013;24:466–480. - "VSports手机版" PubMed
-
- Chen Y, Lemire M, Choufani S, Butcher DT, Grafodatskaya D, Zanke BW, Gallinger S, Hudson TJ, Weksberg R. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics. 2013;8:203–209. - PMC (VSports手机版) - PubMed
Publication types
- VSports注册入口 - Actions
MeSH terms
- Actions (VSports在线直播)
- "V体育官网入口" Actions
- V体育官网 - Actions
- Actions (V体育平台登录)
- Actions (VSports)
- Actions (VSports)
- V体育平台登录 - Actions
- VSports注册入口 - Actions
- "VSports手机版" Actions
- V体育平台登录 - Actions
- "V体育ios版" Actions
Substances
- Actions (V体育安卓版)
- "VSports" Actions
Grants and funding
LinkOut - more resources
"V体育官网入口" Full Text Sources
"VSports" Other Literature Sources
Medical
Molecular Biology Databases

