Recipient mucosal-associated invariant T cells control GVHD within the colon
- PMID: 29629900
- PMCID: PMC5919881
- DOI: "V体育官网" 10.1172/JCI91646
V体育ios版 - Recipient mucosal-associated invariant T cells control GVHD within the colon
Abstract
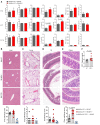
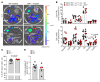
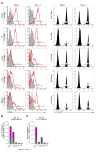
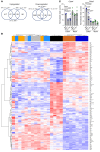
Mucosal-associated invariant T (MAIT) cells are a unique innate-like T cell subset that responds to a wide array of bacteria and yeast through recognition of riboflavin metabolites presented by the MHC class I-like molecule MR1. Here, we demonstrate using MR1 tetramers that recipient MAIT cells are present in small but definable numbers in graft-versus-host disease (GVHD) target organs and protect from acute GVHD in the colon following bone marrow transplantation (BMT). Consistent with their preferential juxtaposition to microbial signals in the colon, recipient MAIT cells generate large amounts of IL-17A, promote gastrointestinal tract integrity, and limit the donor alloantigen presentation that in turn drives donor Th1 and Th17 expansion specifically in the colon after BMT. Allogeneic BMT recipients deficient in IL-17A also develop accelerated GVHD, suggesting MAIT cells likely regulate GVHD, at least in part, by the generation of this cytokine. Indeed, analysis of stool microbiota and colon tissue from IL-17A-/- and MR1-/- mice identified analogous shifts in microbiome operational taxonomic units (OTU) and mediators of barrier integrity that appear to represent pathways controlled by similar, IL-17A-dependent mechanisms. Thus, MAIT cells act to control barrier function to attenuate pathogenic T cell responses in the colon and, given their very high frequency in humans, likely represent an important population in clinical BMT VSports手机版. .
Keywords: Bone marrow transplantation; Immunology; Transplantation. V体育安卓版.
Conflict of interest statement (V体育ios版)
Conflict of interest: LKN, JR, JM, and DPF are named inventors on a patent application (PCT/AU2013/000742, WO2014005194) V体育ios版. LKN, JYWM, JR, JM, and DPF are named inventors on a patent application (PCT/AU2015/050148, WO2015149130) involving MR1 ligands for MR1-restricted MAIT cells owned by University of Queensland, Monash University, and University of Melbourne.
Figures






"VSports在线直播" References
-
- Hill GR, Ferrara JL. The primacy of the gastrointestinal tract as a target organ of acute graft-versus-host disease: rationale for the use of cytokine shields in allogeneic bone marrow transplantation. Blood. 2000;95(9):2754–2759. - PubMed
-
- Cua DJ, Tato CM. Innate IL-17-producing cells: the sentinels of the immune system. Nat Rev Immunol. 2010;10(7):479–489. doi: 10.1038/nri2800. - DOI (V体育官网) - PubMed
Publication types
MeSH terms
- V体育官网 - Actions
- V体育安卓版 - Actions
- Actions (VSports注册入口)
- Actions (V体育ios版)
- VSports手机版 - Actions
- V体育2025版 - Actions
- "V体育2025版" Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

