V体育官网入口 - Cand1-Mediated Adaptive Exchange Mechanism Enables Variation in F-Box Protein Expression
- PMID: 29499133
- PMCID: "V体育ios版" PMC5836512
- DOI: 10.1016/j.molcel.2018.01.038
"VSports手机版" Cand1-Mediated Adaptive Exchange Mechanism Enables Variation in F-Box Protein Expression
Abstract (VSports手机版)
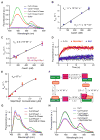
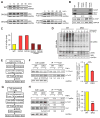
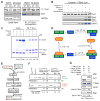
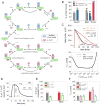
Skp1⋅Cul1⋅F-box (SCF) ubiquitin ligase assembly is regulated by the interplay of substrate binding, reversible Nedd8 conjugation on Cul1, and the F-box protein (FBP) exchange factors Cand1 and Cand2. Detailed investigations into SCF assembly and function in reconstituted systems and Cand1/2 knockout cells informed the development of a mathematical model for how dynamical assembly of SCF complexes is controlled and how this cycle is coupled to degradation of an SCF substrate. Simulations predicted an unanticipated hypersensitivity of Cand1/2-deficient cells to FBP expression levels, which was experimentally validated VSports手机版. Together, these and prior observations lead us to propose the adaptive exchange hypothesis, which posits that regulation of the koff of an FBP from SCF by the actions of substrate, Nedd8, and Cand1 molds the cellular repertoire of SCF complexes and that the plasticity afforded by this exchange mechanism may enable large variations in FBP expression during development and in FBP gene number during evolution. .
Keywords: Cand1; SCF cycle; exchange factor; mathematical modeling; ubiquitin ligase V体育安卓版. .
Copyright © 2018 Elsevier Inc. All rights reserved V体育ios版. .
Conflict of interest statement
Raymond J. Deshaies is an employee and shareholder of Amgen VSports最新版本.
VSports在线直播 - Figures

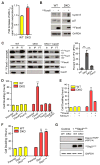
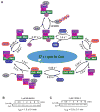
Comment in
-
SCF E3 Ligase Substrates Switch from CAN-D to Can-ubiquitylate.Mol Cell. 2018 Mar 1;69(5):721-723. doi: 10.1016/j.molcel.2018.02.019. Mol Cell. 2018. PMID: 29499128
References
-
- Bosu DR, Feng H, Min K, Kim Y, Wallenfang MR, Kipreos ET. C. elegans CAND-1 regulates cullin neddylation, cell proliferation and morphogenesis in specific tissues. Developmental biology. 2010;346:113–126. - "VSports手机版" PMC - PubMed
Publication types
V体育平台登录 - MeSH terms
- Actions (VSports最新版本)
- VSports - Actions
- V体育安卓版 - Actions
- "V体育官网" Actions
- "V体育官网入口" Actions
- "VSports最新版本" Actions
VSports在线直播 - Substances
- Actions (V体育官网入口)
- V体育安卓版 - Actions
- "VSports" Actions
Grants and funding
LinkOut - more resources
V体育官网入口 - Full Text Sources
Other Literature Sources
VSports在线直播 - Molecular Biology Databases
Miscellaneous

