VSports app下载 - Dynamic Regulation of Long-Chain Fatty Acid Oxidation by a Noncanonical Interaction between the MCL-1 BH3 Helix and VLCAD
- PMID: 29499131
- PMCID: PMC5916823
- DOI: 10.1016/j.molcel.2018.02.005
Dynamic Regulation of Long-Chain Fatty Acid Oxidation by a Noncanonical Interaction between the MCL-1 BH3 Helix and VLCAD
Abstract (V体育2025版)
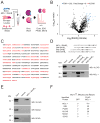
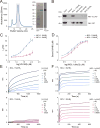
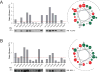
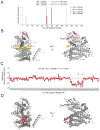
MCL-1 is a BCL-2 family protein implicated in the development and chemoresistance of human cancer VSports手机版. Unlike its anti-apoptotic homologs, Mcl-1 deletion has profound physiologic consequences, indicative of a broader role in homeostasis. We report that the BCL-2 homology 3 (BH3) α helix of MCL-1 can directly engage very long-chain acyl-CoA dehydrogenase (VLCAD), a key enzyme of the mitochondrial fatty acid β-oxidation (FAO) pathway. Proteomic analysis confirmed that the mitochondrial matrix isoform of MCL-1 (MCL-1Matrix) interacts with VLCAD. Mcl-1 deletion, or eliminating MCL-1Matrix alone, selectively deregulated long-chain FAO, causing increased flux through the pathway in response to nutrient deprivation. Transient elevation in MCL-1 upon serum withdrawal, a striking increase in MCL-1 BH3/VLCAD interaction upon palmitic acid titration, and direct modulation of enzymatic activity by the MCL-1 BH3 α helix are consistent with dynamic regulation. Thus, the MCL-1 BH3 interaction with VLCAD revealed a separable, gain-of-function role for MCL-1 in the regulation of lipid metabolism. .
Keywords: BCL-2 family; MCL-1; VLCAD; apoptosis; fatty acid metabolism; mitochondria; mitochondrial matrix; stapled peptide; α helix; β-oxidation V体育安卓版. .
Copyright © 2018 Elsevier Inc. All rights reserved V体育ios版. .
Conflict of interest statement
L. D. W. is a scientific advisory board member and consultant for Aileron Therapeutics VSports最新版本. L. D. W. and S. E. have a patent filing related to this work.
Figures
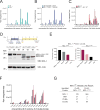

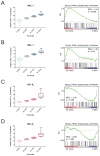
References
-
- Aouacheria A, Combet C, Tompa P, Hardwick JM. Redefining the BH3 Death Domain as a ‘Short Linear Motif’. Trends Biochem Sci. 2015;40:736–748. - VSports最新版本 - PMC - PubMed
-
- Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. - PMC (VSports app下载) - PubMed
Publication types
- V体育官网入口 - Actions
- "V体育ios版" Actions
MeSH terms
- VSports - Actions
- Actions (V体育ios版)
- "VSports app下载" Actions
- "V体育平台登录" Actions
- "V体育安卓版" Actions
- V体育安卓版 - Actions
- Actions (V体育2025版)
- "V体育ios版" Actions
Substances
- V体育ios版 - Actions
- "V体育2025版" Actions
Grants and funding
- R01 CA166429/CA/NCI NIH HHS/United States
- "V体育安卓版" DFS-134963/CIHR/Canada
- P30 DK034854/DK/NIDDK NIH HHS/United States
- P30 CA021765/CA/NCI NIH HHS/United States
- V体育2025版 - T32 GM008313/GM/NIGMS NIH HHS/United States
- V体育2025版 - R35 CA197583/CA/NCI NIH HHS/United States
- R50 CA211399/CA/NCI NIH HHS/United States
- R01 HL102175/HL/NHLBI NIH HHS/United States
- R01 CA201069/CA/NCI NIH HHS/United States
- R01 CA217092/CA/NCI NIH HHS/United States
- R01 HL123543/HL/NHLBI NIH HHS/United States
- T32 GM007753/GM/NIGMS NIH HHS/United States
- R01 DK103295/DK/NIDDK NIH HHS/United States
LinkOut - more resources
V体育平台登录 - Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

