Targeting IRF3 as a YAP agonist therapy against gastric cancer
- PMID: 29339449
- PMCID: PMC5789414
- DOI: 10.1084/jem.20171116 (VSports最新版本)
Targeting IRF3 as a YAP agonist therapy against gastric cancer
Abstract
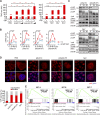
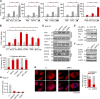
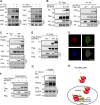
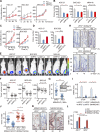
The Hippo pathway plays a vital role in tissue homeostasis and tumorigenesis. The transcription factor IRF3 is essential for innate antiviral immunity. In this study, we discovered IRF3 as an agonist of Yes-associated protein (YAP). The expression of IRF3 is positively correlated with that of YAP and its target genes in gastric cancer; the expression of both IRF3 and YAP is up-regulated and prognosticates patient survival. IRF3 interacts with both YAP and TEAD4 in the nucleus to enhance their interaction, promoting nuclear translocation and activation of YAP. IRF3 and YAP-TEAD4 are associated genome-wide to cobind and coregulate many target genes of the Hippo pathway. Overexpression of active IRF3 increased, but depletion of IRF3 reduced, the occupancy of YAP on the target genes. Knockdown or pharmacological targeting of IRF3 by Amlexanox, a drug used clinically for antiinflammatory treatment, inhibits gastric tumor growth in a YAP-dependent manner VSports手机版. Collectively, our study identifies IRF3 as a positive regulator for YAP, highlighting a new therapeutic target against YAP-driven cancers. .
© 2018 Jiao et al.
Figures



References
-
- Bahrami A., Amerizadeh F., ShahidSales S., Khazaei M., Ghayour-Mobarhan M., Sadeghnia H.R., Maftouh M., Hassanian S.M., and Avan A.. 2017. Therapeutic potential of targeting Wnt/β-catenin pathway in treatment of colorectal cancer: Rational and progress. J. Cell. Biochem. 118:1979–1983. 10.1002/jcb.25903 - VSports最新版本 - DOI - PubMed
-
- Cai J., Zhang N., Zheng Y., de Wilde R.F., Maitra A., and Pan D.. 2010. The Hippo signaling pathway restricts the oncogenic potential of an intestinal regeneration program. Genes Dev. 24:2383–2388. 10.1101/gad.1978810 - DOI (V体育安卓版) - PMC - PubMed
-
- Chan S.W., Lim C.J., Chen L., Chong Y.F., Huang C., Song H., and Hong W.. 2011. The Hippo pathway in biological control and cancer development. J. Cell. Physiol. 226:928–939. 10.1002/jcp.22435 - V体育ios版 - DOI - PubMed
Publication types
- "VSports注册入口" Actions
MeSH terms
- Actions (V体育平台登录)
- Actions (V体育平台登录)
- Actions (V体育官网入口)
- V体育平台登录 - Actions
- "VSports在线直播" Actions
- V体育2025版 - Actions
- "VSports注册入口" Actions
- "VSports" Actions
- VSports - Actions
- V体育ios版 - Actions
- Actions (VSports注册入口)
- Actions (V体育安卓版)
- VSports注册入口 - Actions
- "VSports最新版本" Actions
- VSports最新版本 - Actions
- VSports注册入口 - Actions
V体育平台登录 - Substances
- Actions (VSports手机版)
- V体育2025版 - Actions
- V体育ios版 - Actions
- V体育安卓版 - Actions
- "V体育平台登录" Actions
- "VSports app下载" Actions
- Actions (VSports最新版本)
- V体育ios版 - Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
"VSports app下载" Medical
V体育官网 - Molecular Biology Databases
Research Materials

