Improved methods for predicting peptide binding affinity to MHC class II molecules
- PMID: 29315598
- PMCID: PMC6002223
- DOI: V体育安卓版 - 10.1111/imm.12889
Improved methods for predicting peptide binding affinity to MHC class II molecules
Abstract
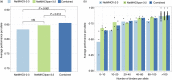
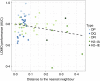
Major histocompatibility complex class II (MHC-II) molecules are expressed on the surface of professional antigen-presenting cells where they display peptides to T helper cells, which orchestrate the onset and outcome of many host immune responses. Understanding which peptides will be presented by the MHC-II molecule is therefore important for understanding the activation of T helper cells and can be used to identify T-cell epitopes. We here present updated versions of two MHC-II-peptide binding affinity prediction methods, NetMHCII and NetMHCIIpan. These were constructed using an extended data set of quantitative MHC-peptide binding affinity data obtained from the Immune Epitope Database covering HLA-DR, HLA-DQ, HLA-DP and H-2 mouse molecules. We show that training with this extended data set improved the performance for peptide binding predictions for both methods. Both methods are publicly available at www. cbs. dtu. dk/services/NetMHCII-2 VSports手机版. 3 and www. cbs. dtu. dk/services/NetMHCIIpan-3. 2. .
Keywords: MHC binding specificity; T-cell epitope; affinity predictions; immunogenic peptides; peptide-MHC binding V体育安卓版. .
© 2018 John Wiley & Sons Ltd.
Figures (V体育平台登录)


References
-
- Castellino F, Zhong G, Germain RN. Antigen presentation by MHC class II molecules: invariant chain function, protein trafficking, and the molecular basis of diverse determinant capture. Hum Immunol 1997; 54:159–69. - PubMed
-
- Lin HH, Zhang GL, Tongchusak S, Reinherz EL, Brusic V. Evaluation of MHC‐II peptide binding prediction servers: applications for vaccine research. BMC Bioinformatics 2008; 9(Suppl. 12):S22. - PMC (V体育平台登录) - PubMed
-
- Nielsen M, Lund O, Buus S, Lundegaard C. MHC Class II epitope predictive algorithms. Immunology 2010; 130:319–28. - PMC (VSports app下载) - PubMed
-
- Brown JH, Jardetzky TS, Gorga JC, Stern LJ, Urban RG, Strominger JL et al Three‐dimensional structure of the human Class II histocompatibility antigen HLA‐DR1. J Immunol 2015; 194:5–11. - "V体育2025版" PubMed
Publication types
- Actions (VSports注册入口)
MeSH terms
- "V体育ios版" Actions
- VSports注册入口 - Actions
- Actions (V体育ios版)
- V体育安卓版 - Actions
- V体育2025版 - Actions
- VSports最新版本 - Actions
- "VSports最新版本" Actions
- "V体育ios版" Actions
Substances
- Actions (VSports最新版本)
"V体育ios版" Grants and funding
LinkOut - more resources
"VSports app下载" Full Text Sources
Other Literature Sources
Research Materials

