MYC Drives a Subset of High-Risk Pediatric Neuroblastomas and Is Activated through Mechanisms Including Enhancer Hijacking and Focal Enhancer Amplification (V体育官网)
- PMID: 29284669
- PMCID: VSports最新版本 - PMC5856009
- DOI: 10.1158/2159-8290.CD-17-0993
MYC Drives a Subset of High-Risk Pediatric Neuroblastomas and Is Activated through Mechanisms Including Enhancer Hijacking and Focal Enhancer Amplification (V体育平台登录)
Abstract
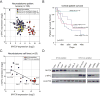
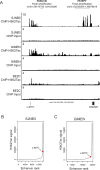
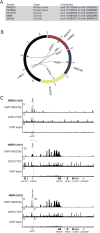
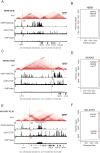
The amplified MYCN gene serves as an oncogenic driver in approximately 20% of high-risk pediatric neuroblastomas. Here, we show that the family member MYC is a potent transforming gene in a separate subset of high-risk neuroblastoma cases (∼10%), based on (i) its upregulation by focal enhancer amplification or genomic rearrangements leading to enhancer hijacking, and (ii) its ability to transform neuroblastoma precursor cells in a transgenic animal model. The aberrant regulatory elements associated with oncogenic MYC activation include focally amplified distal enhancers and translocation of highly active enhancers from other genes to within topologically associating domains containing the MYC gene locus. The clinical outcome for patients with high levels of MYC expression is virtually identical to that of patients with amplification of the MYCN gene, a known high-risk feature of this disease. Together, these findings establish MYC as a bona fide oncogene in a clinically significant group of high-risk childhood neuroblastomas. Significance: Amplification of the MYCN oncogene is a recognized hallmark of high-risk pediatric neuroblastoma. Here, we demonstrate that MYC is also activated as a potent oncogene in a distinct subset of neuroblastoma cases through either focal amplification of distal enhancers or enhancer hijacking mediated by chromosomal translocation. Cancer Discov; 8(3); 320-35. ©2017 AACR VSports手机版. This article is highlighted in the In This Issue feature, p. 253. .
©2017 American Association for Cancer Research. V体育安卓版.
Conflict of interest statement
The authors do have any conflicts of interest to disclose.
"V体育ios版" Figures

References (VSports在线直播)
-
- Matthay KK, Maris JM, Schleiermacher G, Nakagawara A, Mackall CL, Diller L, et al. Neuroblastoma. Nat Rev Dis Primers. 2016;16078:2. - PubMed (VSports手机版)
-
- Brodeur GM, RC S, M S, HE V, JM B. Amplification of N-myc in untreated human neuroblastomas correlates with advanced disease stage. Science. 1984;224:1121–4. - PubMed
-
- Wang LL, Teshiba R, Ikegaki N, Tang XX, Naranjo A, London WB, et al. Augmented expression of MYC and/or MYCN protein defines highly aggressive MYC-driven neuroblastoma: a Children’s Oncology Group study. Br J Cancer. 2015;113:57–63. - "V体育安卓版" PMC - PubMed
-
- Westermann F, Muth D, Benner A, Bauer T, Henrich KO, Oberthuer A, et al. Distinct transcriptional MYCN/c-MYC activities are associated with spontaneous regression or malignant progression in neuroblastomas. Genome Biol. 2008;9:R150. - PMC (V体育2025版) - PubMed
Publication types
- "VSports" Actions
MeSH terms
- "V体育安卓版" Actions
- "VSports在线直播" Actions
- Actions (V体育官网入口)
- "V体育2025版" Actions
- Actions (VSports在线直播)
- "VSports手机版" Actions
- Actions (VSports手机版)
- "VSports最新版本" Actions
- Actions (VSports手机版)
- "V体育官网入口" Actions
- "V体育平台登录" Actions
- V体育安卓版 - Actions
- VSports注册入口 - Actions
Substances
- "VSports注册入口" Actions
- Actions (VSports注册入口)
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
V体育安卓版 - Molecular Biology Databases

