Distinctive roles of age, sex, and genetics in shaping transcriptional variation of human immune responses to microbial challenges
- PMID: 29282317
- PMCID: "V体育官网入口" PMC5776984
- DOI: 10.1073/pnas.1714765115
"VSports app下载" Distinctive roles of age, sex, and genetics in shaping transcriptional variation of human immune responses to microbial challenges
Abstract
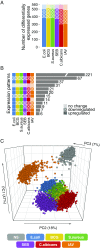
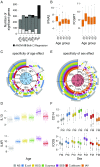
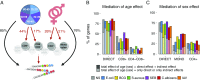
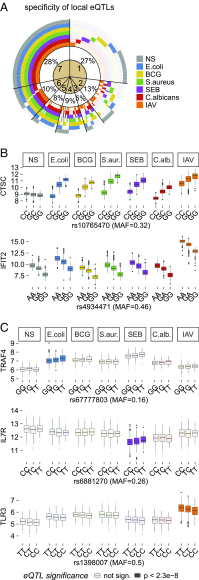
The contribution of host genetic and nongenetic factors to immunological differences in humans remains largely undefined. Here, we generated bacterial-, fungal-, and viral-induced immune transcriptional profiles in an age- and sex-balanced cohort of 1,000 healthy individuals and searched for the determinants of immune response variation. We found that age and sex affected the transcriptional response of most immune-related genes, with age effects being more stimulus-specific relative to sex effects, which were largely shared across conditions. Although specific cell populations mediated the effects of age and sex on gene expression, including CD8+ T cells for age and CD4+ T cells and monocytes for sex, we detected a direct effect of these intrinsic factors for the majority of immune genes VSports手机版. The mapping of expression quantitative trait loci (eQTLs) revealed that genetic factors had a stronger effect on immune gene regulation than age and sex, yet they affected a smaller number of genes. Importantly, we identified numerous genetic variants that manifested their regulatory effects exclusively on immune stimulation, including a Candida albicans-specific master regulator at the CR1 locus. These response eQTLs were enriched in disease-associated variants, particularly for autoimmune and inflammatory disorders, indicating that differences in disease risk may result from regulatory variants exerting their effects only in the presence of immune stress. Together, this study quantifies the respective effects of age, sex, genetics, and cellular heterogeneity on the interindividual variability of immune responses and constitutes a valuable resource for further exploration in the context of different infection risks or disease outcomes. .
Keywords: age; gene expression; genetics; human immune variation; sex. V体育安卓版.
Copyright © 2018 the Author(s). Published by PNAS. V体育ios版.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- De Jager PL, et al. ImmVar project: Insights and design considerations for future studies of “healthy” immune variation. Semin Immunol. 2015;27:51–57. - PubMed
-
- Liston A, Carr EJ, Linterman MA. Shaping variation in the human immune system. Trends Immunol. 2016;37:637–646. - PubMed
-
- Davis MM, Tato CM, Furman D. Systems immunology: Just getting started. Nat Immunol. 2017;18:725–732. - "VSports在线直播" PMC - PubMed
-
- Brodin P, et al. Variation in the human immune system is largely driven by non-heritable influences. Cell. 2015;160:37–47. - PMC (VSports手机版) - PubMed
Publication types
- Actions (V体育ios版)
MeSH terms
- Actions (VSports注册入口)
- Actions (VSports注册入口)
Substances
- "V体育安卓版" Actions
"V体育ios版" LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

