"V体育平台登录" The gut mycobiome of the Human Microbiome Project healthy cohort
- PMID: 29178920
- PMCID: PMC5702186
- DOI: 10.1186/s40168-017-0373-4
The gut mycobiome of the Human Microbiome Project healthy cohort
Abstract
Background: Most studies describing the human gut microbiome in healthy and diseased states have emphasized the bacterial component, but the fungal microbiome (i. e. , the mycobiome) is beginning to gain recognition as a fundamental part of our microbiome. To date, human gut mycobiome studies have primarily been disease centric or in small cohorts of healthy individuals. To contribute to existing knowledge of the human mycobiome, we investigated the gut mycobiome of the Human Microbiome Project (HMP) cohort by sequencing the Internal Transcribed Spacer 2 (ITS2) region as well as the 18S rRNA gene VSports手机版. .
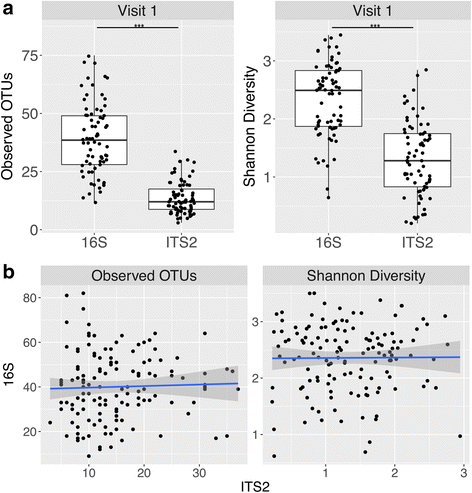
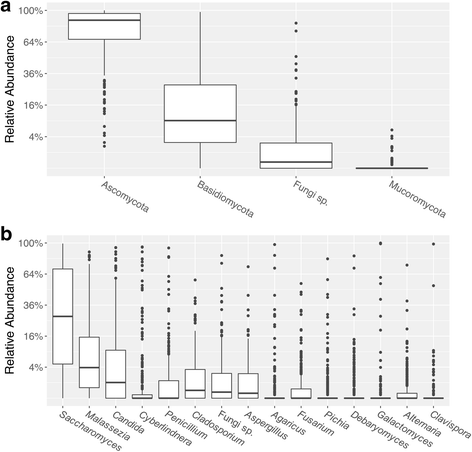
Results: Three hundred seventeen HMP stool samples were analyzed by ITS2 sequencing. Fecal fungal diversity was significantly lower in comparison to bacterial diversity. Yeast dominated the samples, comprising eight of the top 15 most abundant genera. Specifically, fungal communities were characterized by a high prevalence of Saccharomyces, Malassezia, and Candida, with S. cerevisiae, M. restricta, and C. albicans operational taxonomic units (OTUs) present in 96. 8, 88. 3, and 80. 8% of samples, respectively. There was a high degree of inter- and intra-volunteer variability in fungal communities. However, S. cerevisiae, M. restricta, and C. albicans OTUs were found in 92. 2, 78. 3, and 63. 6% of volunteers, respectively, in all samples donated over an approximately 1-year period. Metagenomic and 18S rRNA gene sequencing data agreed with ITS2 results; however, ITS2 sequencing provided greater resolution of the relatively low abundance mycobiome constituents. V体育安卓版.
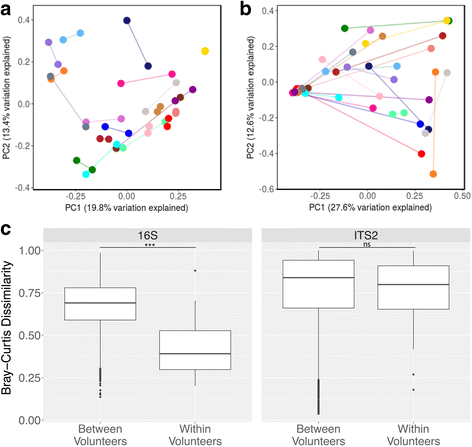
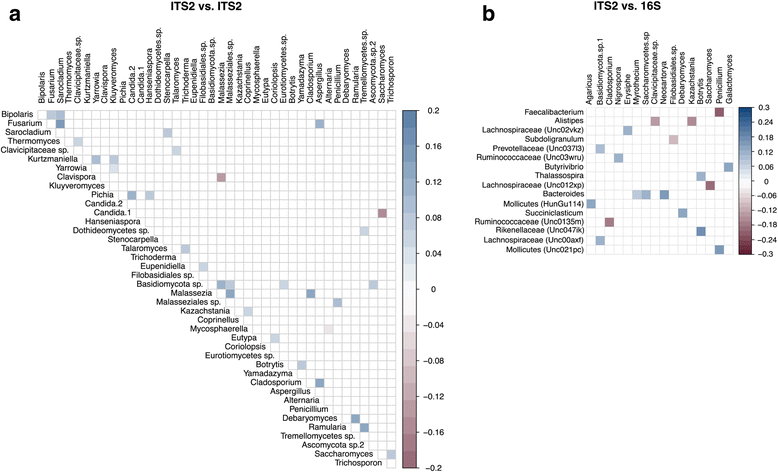
Conclusions: Compared to bacterial communities, the human gut mycobiome is low in diversity and dominated by yeast including Saccharomyces, Malassezia, and Candida V体育ios版. Both inter- and intra-volunteer variability in the HMP cohort were high, revealing that unlike bacterial communities, an individual's mycobiome is no more similar to itself over time than to another person's. Nonetheless, several fungal species persisted across a majority of samples, evidence that a core gut mycobiome may exist. ITS2 sequencing data provided greater resolution of the mycobiome membership compared to metagenomic and 18S rRNA gene sequencing data, suggesting that it is a more sensitive method for studying the mycobiome of stool samples. .
Keywords: Fecal microbiome; Fungal microbiome; Fungi; HMP; ITS2; Metagenomics; Microbiome; Microbiota; There was a high degree o Spacer VSports最新版本. .
Conflict of interest statement (VSports app下载)
Ethics approval and consent to participate
The Institutional Review Board (IRB) at Baylor College of Medicine reviewed and approved the protocols, informed consent, and other study documents (IRB protocols H-22895 (IRB no. 00001021) and H-22035 (IRB no V体育平台登录. 00002649)) [16].
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
"VSports" Figures
References
-
- Huffnagle GB, Noverr MC. The emerging world of the fungal microbiome. Trends Microbiol. 2013;21:334–341. doi: 10.1016/j.tim.2013.04.002. - DOI (V体育ios版) - PMC - PubMed
MeSH terms
- "VSports" Actions
- "VSports注册入口" Actions
- "V体育ios版" Actions
- Actions (VSports)
- VSports最新版本 - Actions
- "V体育ios版" Actions
- "VSports手机版" Actions
- "VSports" Actions
- Actions (VSports)
- V体育平台登录 - Actions
- "V体育平台登录" Actions
- VSports最新版本 - Actions
- "VSports注册入口" Actions
- VSports手机版 - Actions
- "V体育安卓版" Actions
- Actions (V体育2025版)
Substances (VSports)
Grants and funding
LinkOut - more resources (VSports在线直播)
V体育安卓版 - Full Text Sources
V体育2025版 - Other Literature Sources
Medical
Molecular Biology Databases

