Targeting the MYCN-PARP-DNA Damage Response Pathway in Neuroendocrine Prostate Cancer (V体育平台登录)
- PMID: 29138344
- PMCID: PMC5823274
- DOI: VSports注册入口 - 10.1158/1078-0432.CCR-17-1872
Targeting the MYCN-PARP-DNA Damage Response Pathway in Neuroendocrine Prostate Cancer
Abstract
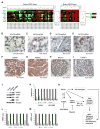
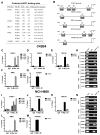
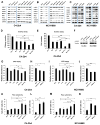
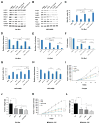
Purpose: We investigated MYCN-regulated molecular pathways in castration-resistant prostate cancer (CRPC) classified by morphologic criteria as adenocarcinoma or neuroendocrine to extend the molecular phenotype, establish driver pathways, and identify novel approaches to combination therapy for neuroendocrine prostate cancer (NEPC). Experimental Design and Results: Using comparative bioinformatics analyses of CRPC-Adeno and CRPC-Neuro RNA sequence data from public data sets and a panel of 28 PDX models, we identified a MYCN-PARP-DNA damage response (DDR) pathway that is enriched in CRPC with neuroendocrine differentiation (NED) and CRPC-Neuro. ChIP-PCR assay revealed that N-MYC transcriptionally activates PARP1, PARP2, BRCA1, RMI2, and TOPBP1 through binding to the promoters of these genes. MYCN or PARP1 gene knockdown significantly reduced the expression of MYCN-PARP-DDR pathway genes and NED markers, and inhibition with MYCNsi and/or PARPsi, BRCA1si, or RMI2si significantly suppressed malignant activities, including cell viability, colony formation, and cell migration, in C4-2b4 and NCI-H660 cells. Targeting this pathway with AURKA inhibitor PHA739358 and PARP inhibitor olaparib generated therapeutic effects similar to those of gene knockdown in vitro and significantly suppressed tumor growth in both C4-2b4 and MDACC PDX144-13C subcutaneous models in vivoConclusions: Our results identify a novel MYCN-PARP-DDR pathway that is driven by N-MYC in a subset of CRPC-Adeno and in NEPC. Targeting this pathway using in vitro and in vivo CRPC-Adeno and CRPC-Neuro models demonstrated a novel therapeutic strategy for NEPC. Further investigation of N-MYC-regulated DDR gene targets and the biological and clinical significance of MYCN-PARP-DDR signaling will more fully elucidate the importance of the MYCN-PARP-DDR signaling pathway in the development and maintenance of NEPC. Clin Cancer Res; 24(3); 696-707 VSports手机版. ©2017 AACR. .
©2017 American Association for Cancer Research V体育安卓版. .
V体育官网入口 - Conflict of interest statement
"VSports注册入口" Figures

V体育安卓版 - Comment in
-
Prostate cancer: Damaged good - targeting DNA damage repair in neuroendocrine disease.Nat Rev Urol. 2018 Feb;15(2):70. doi: 10.1038/nrurol.2017.206. Epub 2017 Dec 12. Nat Rev Urol. 2018. PMID: 29231196 No abstract available.
References
-
- Wang HT, Yao YH, Li BG, Tang Y, Chang JW, Zhang J. Neuroendocrine Prostate Cancer (NEPC) progressing from conventional prostatic adenocarcinoma: factors associated with time to development of NEPC and survival from NEPC diagnosis-a systematic review and pooled analysis. J Clin Oncol. 2014;32:3383–90. - PubMed
-
- Helpap B, Kollermann J, Oehler U. Neuroendocrine differentiation in prostatic carcinomas: histogenesis, biology, clinical relevance, and future therapeutical perspectives. Urol Int. 1999;62:133–8. - PubMed
-
- Palmgren JS, Karavadia SS, Wakefield MR. Unusual and underappreciated: small cell carcinoma of the prostate. Semin Oncol. 2007;34:22–9. - PubMed
-
- Spiess PE, Pettaway CA, Vakar-Lopez F, Kassouf W, Wang X, Busby JE, et al. Treatment outcomes of small cell carcinoma of the prostate: a single-center study. Cancer. 2007;110:1729–37. - PubMed (V体育官网)
"V体育安卓版" Publication types
- VSports最新版本 - Actions
MeSH terms (V体育ios版)
- VSports手机版 - Actions
- "V体育平台登录" Actions
- Actions (V体育官网入口)
- V体育安卓版 - Actions
- V体育ios版 - Actions
- "VSports app下载" Actions
- "V体育2025版" Actions
- "V体育安卓版" Actions
- "V体育官网" Actions
- VSports注册入口 - Actions
- "V体育官网" Actions
- Actions (V体育2025版)
- "VSports" Actions
- "VSports app下载" Actions
- Actions (V体育ios版)
- "VSports" Actions
- "V体育安卓版" Actions
- Actions (V体育ios版)
- V体育2025版 - Actions
Substances
- "VSports手机版" Actions
- Actions (V体育官网)
- "VSports在线直播" Actions
- Actions (VSports手机版)
Grants and funding
LinkOut - more resources
Full Text Sources
"V体育ios版" Other Literature Sources
"V体育官网" Medical
Miscellaneous

