Resistin-like molecule β is a bactericidal protein that promotes spatial segregation of the microbiota and the colonic epithelium
- PMID: 28973871
- PMCID: PMC5651776
- DOI: 10.1073/pnas.1711395114
Resistin-like molecule β is a bactericidal protein that promotes spatial segregation of the microbiota and the colonic epithelium
Abstract
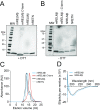
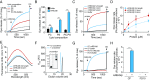
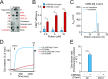
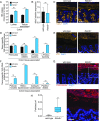
The mammalian intestine is colonized by trillions of bacteria that perform essential metabolic functions for their hosts. The mutualistic nature of this relationship depends on maintaining spatial segregation between these bacteria and the intestinal epithelial surface. This segregation is achieved in part by the presence of a dense mucus layer at the epithelial surface and by the production of antimicrobial proteins that are secreted by epithelial cells into the mucus layer VSports手机版. Here, we show that resistin-like molecule β (RELMβ) is a bactericidal protein that limits contact between Gram-negative bacteria and the colonic epithelial surface. Mouse and human RELMβ selectively killed Gram-negative bacteria by forming size-selective pores that permeabilized bacterial membranes. In mice lacking RELMβ, Proteobacteria were present in the inner mucus layer and invaded mucosal tissues. Another RELM family member, human resistin, was also bactericidal, suggesting that bactericidal activity is a conserved function of the RELM family. Our findings thus identify the RELM family as a unique family of bactericidal proteins and show that RELMβ promotes host-bacterial mutualism by regulating the spatial segregation between the microbiota and the intestinal epithelium. .
Keywords: antibacterial protein; innate immunity; intestinal epithelium; microbiota. V体育安卓版.
Conflict of interest statement
The authors declare no conflict of interest.
Figures (V体育2025版)






"V体育平台登录" References
-
- Okumura R, et al. Lypd8 promotes the segregation of flagellated microbiota and colonic epithelia. Nature. 2016;532:117–121. - PubMed
Publication types
- VSports手机版 - Actions
MeSH terms
- Actions (V体育ios版)
- VSports app下载 - Actions
- "V体育ios版" Actions
- "V体育ios版" Actions
- "VSports在线直播" Actions
Substances
- V体育2025版 - Actions
Grants and funding
LinkOut - more resources
Full Text Sources (V体育官网入口)
Other Literature Sources
Molecular Biology Databases (VSports手机版)
Research Materials (VSports注册入口)

