Novel Role of FBXW7 Circular RNA in Repressing Glioma Tumorigenesis (V体育2025版)
- PMID: 28903484
- PMCID: PMC6019044
- DOI: "V体育ios版" 10.1093/jnci/djx166
Novel Role of FBXW7 Circular RNA in Repressing Glioma Tumorigenesis
Erratum in
-
Corrigendum. (VSports在线直播)J Natl Cancer Inst. 2018 Oct 1;110(10):1147. doi: 10.1093/jnci/djy118. J Natl Cancer Inst. 2018. PMID: 29986068 Free PMC article. No abstract available.
Abstract
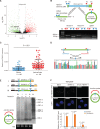
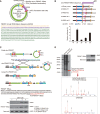
Background: Circular RNAs (circRNAs) are RNA transcripts that are widespread in the eukaryotic genome. Recent evidence indicates that circRNAs play important roles in tissue development, gene regulation, and carcinogenesis VSports手机版. However, whether circRNAs encode functional proteins remains elusive, although translation of several circRNAs was recently reported. .
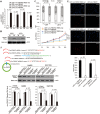
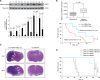
Methods: CircRNA deep sequencing was performed by using 10 pathologically diagnosed glioblastoma samples and their paired adjacent normal brain tissues V体育安卓版. Northern blotting, Sanger sequencing, antibody, and liquid chromatograph Tandem Mass Spectrometer were used to confirm the existence of circ-FBXW7 and its encoded protein in in two cell lines. Lentivirus-transfected stable U251 and U373 cells were used to assess the biological functions of the novel protein invitro and invivo (five mice per group). Clinical implications of circ-FBXW7 were assessed in 38 pathologically diagnosed glioblastoma samples and their paired periphery normal brain tissues by using quantitative polymerase chain reaction (two-sided log-rank test). .
Results: Circ-FBXW7 is abundantly expressed in the normal human brain (reads per kilobase per million mapped reads [RPKM] = 9. 31). The spanning junction open reading frame in circ-FBXW7 driven by internal ribosome entry site encodes a novel 21-kDa protein, which we termed FBXW7-185aa. Upregulation of FBXW7-185aa in cancer cells inhibited proliferation and cell cycle acceleration, while knockdown of FBXW7-185aa promoted malignant phenotypes invitro and invivo. FBXW7-185aa reduced the half-life of c-Myc by antagonizing USP28-induced c-Myc stabilization. Moreover, circ-FBXW7 and FBXW7-185aa levels were reduced in glioblastoma clinical samples compared with their paired tumor-adjacent tissues (P < V体育ios版. 001). Circ-FBXW7 expression positively associated with glioblastoma patient overall survival (P = . 03). .
Conclusions: Endogenous circRNA encodes a functional protein in human cells, and circ-FBXW7 and FBXW7-185aa have potential prognostic implications in brain cancer VSports最新版本. .
© The Author 2017. Published by Oxford University Press V体育平台登录. .
Figures



Comment in
-
RE: Novel Role of FBXW7 Circular RNA in Repressing Glioma Tumorigenesis.J Natl Cancer Inst. 2019 Apr 1;111(4):435. doi: 10.1093/jnci/djy116. J Natl Cancer Inst. 2019. PMID: 29986033 No abstract available.
-
Response to H. Chen, Y. Liu, P. Li, and D. Zhu.J Natl Cancer Inst. 2019 Apr 1;111(4):436. doi: 10.1093/jnci/djy117. J Natl Cancer Inst. 2019. PMID: 29986049 No abstract available.
"V体育2025版" References
-
- Nigro JM, Cho KR, Fearon ER et al. , Scrambled exons. Cell. 1991;643:607–613. - PubMed (V体育2025版)
-
- Capel B, Swain A, Nicolis S et al. , Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell. 1993;735:1019–1030. - PubMed
MeSH terms
- Actions (V体育平台登录)
- Actions (V体育平台登录)
- "V体育官网" Actions
- Actions (VSports注册入口)
- VSports手机版 - Actions
- V体育ios版 - Actions
- "V体育安卓版" Actions
- Actions (VSports手机版)
- "VSports在线直播" Actions
- Actions (V体育安卓版)
- "VSports在线直播" Actions
- V体育官网 - Actions
- "V体育平台登录" Actions
- VSports最新版本 - Actions
- "VSports手机版" Actions
- Actions (V体育官网)
- "V体育官网" Actions
Substances
- VSports注册入口 - Actions
- V体育官网入口 - Actions
- VSports app下载 - Actions
- VSports最新版本 - Actions
- Actions (V体育平台登录)
- V体育安卓版 - Actions
- Actions (VSports在线直播)
Grants and funding
LinkOut - more resources
V体育平台登录 - Full Text Sources
Other Literature Sources (V体育官网入口)
VSports - Medical

