"VSports注册入口" Multiple layers of heterogeneity and subset diversity in human MAIT cell responses to distinct microorganisms and to innate cytokines
- PMID: 28630305
- PMCID: "VSports手机版" PMC5502643
- DOI: 10.1073/pnas.1705759114
V体育官网 - Multiple layers of heterogeneity and subset diversity in human MAIT cell responses to distinct microorganisms and to innate cytokines
Abstract
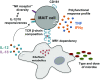
Mucosa-associated invariant T (MAIT) cells are a large innate-like T-cell subset in humans defined by invariant TCR Vα7. 2 use and expression of CD161. MAIT cells recognize microbial riboflavin metabolites of bacterial or fungal origin presented by the monomorphic MR1 molecule. The extraordinary level of evolutionary conservation of MR1 and the limited known diversity of riboflavin metabolite antigens have suggested that MAIT cells are relatively homogeneous and uniform in responses against diverse microbes carrying the riboflavin biosynthesis pathway. The ability of MAIT cells to exhibit microbe-specific functional specialization has not been thoroughly investigated. Here, we found that MAIT cell responses against Escherichia coli and Candida albicans displayed microbe-specific polyfunctional response profiles, antigen sensitivity, and response magnitudes. MAIT cell effector responses against E. coli and C. albicans displayed differential MR1 dependency and TCR β-chain bias, consistent with possible divergent antigen subspecificities between these bacterial and fungal organisms VSports手机版. Finally, although the MAIT cell immunoproteome was overall relatively homogenous and consistent with an effector memory-like profile, it still revealed diversity in a set of natural killer cell-associated receptors. Among these, CD56, CD84, and CD94 defined a subset with higher expression of the transcription factors promyelocytic leukemia zinc finger (PLZF), eomesodermin, and T-bet and enhanced capacity to respond to IL-12 and IL-18 stimulation. Thus, the conserved and innate-like MAIT cells harbor multiple layers of functional heterogeneity as they respond to bacterial or fungal organisms or innate cytokines and adapt their antimicrobial response patterns in a stimulus-specific manner. .
Keywords: MAIT cells; MR1; T cells; immunity; microbial immunity V体育安卓版. .
Conflict of interest statement
The authors declare no conflict of interest.
Figures







References
-
- Dusseaux M, et al. Human MAIT cells are xenobiotic-resistant, tissue-targeted, CD161hi IL-17-secreting T cells. Blood. 2011;117:1250–1259. - PubMed
-
- Treiner E, et al. Selection of evolutionarily conserved mucosal-associated invariant T cells by MR1. Nature. 2003;422:164–169. - PubMed
-
- Lepore M, et al. Parallel T-cell cloning and deep sequencing of human MAIT cells reveal stable oligoclonal TCRβ repertoire. Nat Commun. 2014;5:3866. - PubMed (V体育ios版)
Publication types
MeSH terms
- Actions (V体育安卓版)
- Actions (VSports注册入口)
- Actions (V体育安卓版)
- Actions (V体育官网入口)
- V体育官网入口 - Actions
- VSports手机版 - Actions
- V体育安卓版 - Actions
- Actions (V体育官网)
- Actions (V体育安卓版)
- Actions (VSports手机版)
- "V体育ios版" Actions
Substances
- "V体育官网入口" Actions
Grants and funding
LinkOut - more resources
Full Text Sources (V体育安卓版)
Other Literature Sources
"VSports手机版" Research Materials
Miscellaneous

