A Point Mutation in the Rhesus Rotavirus VP4 Protein Generated through a Rotavirus Reverse Genetics System Attenuates Biliary Atresia in the Murine Model
- PMID: 28515290
- PMCID: PMC5512261
- DOI: VSports手机版 - 10.1128/JVI.00510-17
"V体育安卓版" A Point Mutation in the Rhesus Rotavirus VP4 Protein Generated through a Rotavirus Reverse Genetics System Attenuates Biliary Atresia in the Murine Model
Abstract
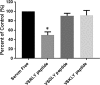
Rotavirus infection is one of the most common causes of diarrheal illness in humans. In neonatal mice, rhesus rotavirus (RRV) can induce biliary atresia (BA), a disease resulting in inflammatory obstruction of the extrahepatic biliary tract and intrahepatic bile ducts. We previously showed that the amino acid arginine (R) within the sequence SRL (amino acids 445 to 447) in the RRV VP4 protein is required for viral binding and entry into biliary epithelial cells. To determine if this single amino acid (R) influences the pathogenicity of the virus, we generated a recombinant virus with a single amino acid mutation at this site through a reverse genetics system VSports手机版. We demonstrated that the RRV mutant (RRVVP4-R446G) produced less symptomatology and replicated to lower titers both in vivo and in vitro than those seen with wild-type RRV, with reduced binding in cholangiocytes. Our results demonstrate that a single amino acid change in the RRV VP4 gene influences cholangiocyte tropism and reduces pathogenicity in mice. IMPORTANCE Rotavirus is the leading cause of diarrhea in humans. Rhesus rotavirus (RRV) can also lead to biliary atresia (a neonatal human disease) in mice. We developed a reverse genetics system to create a mutant of RRV (RRVVP4-R446G) with a single amino acid change in the VP4 protein compared to that of wild-type RRV. In vitro, the mutant virus had reduced binding and infectivity in cholangiocytes. In vivo, it produced fewer symptoms and lower mortality in neonatal mice, resulting in an attenuated form of biliary atresia. .
Keywords: RRV; biliary atresia; cholangiocyte; reverse genetics. V体育安卓版.
Copyright © 2017 American Society for Microbiology. V体育ios版.
"V体育官网入口" Figures


References
-
- Balistreri WF, Grand R, Hoofnagle JH, Suchy FJ, Ryckman FC, Perlmutter DH, Sokol RJ. 1996. Biliary atresia: current concepts and research directions. Summary of a symposium. Hepatology 23:1682–1692. - PubMed
-
- Riepenhoff-Talty M, Gouvea V, Evans MJ, Svensson L, Hoffenberg E, Sokol RJ, Uhnoo I, Greenberg SJ, Schakel K, Zhaori G, Fitzgerald J, Chong S, el-Yousef M, Nemeth A, Brown M, Piccoli D, Hyams J, Ruffin D, Rossi T. 1996. Detection of group C rotavirus in infants with extrahepatic biliary atresia. J Infect Dis 174:8–15. doi:10.1093/infdis/174.1.8. - DOI - PubMed
-
- Glaser JH, Balistreri WF, Morecki R. 1984. Role of reovirus type 3 in persistent infantile cholestasis. J Pediatr 105:912–915. doi:10.1016/S0022-3476(84)80076-1. - "VSports" DOI - PubMed
Publication types
MeSH terms
- "VSports手机版" Actions
- "V体育ios版" Actions
- VSports最新版本 - Actions
- Actions (VSports在线直播)
- Actions (VSports注册入口)
- Actions (VSports注册入口)
- "V体育安卓版" Actions
Substances
- Actions (V体育官网入口)
Grants and funding
LinkOut - more resources
Full Text Sources (VSports)
"V体育2025版" Other Literature Sources
Molecular Biology Databases
Miscellaneous

