Uncovering the trimethylamine-producing bacteria of the human gut microbiota
- PMID: 28506279
- PMCID: PMC5433236
- DOI: 10.1186/s40168-017-0271-9
Uncovering the trimethylamine-producing bacteria of the human gut microbiota
Abstract
Background: Trimethylamine (TMA), produced by the gut microbiota from dietary quaternary amines (mainly choline and carnitine), is associated with atherosclerosis and severe cardiovascular disease. Currently, little information on the composition of TMA producers in the gut is available due to their low abundance and the requirement of specific functional-based detection methods as many taxa show disparate abilities to produce that compound. VSports手机版.
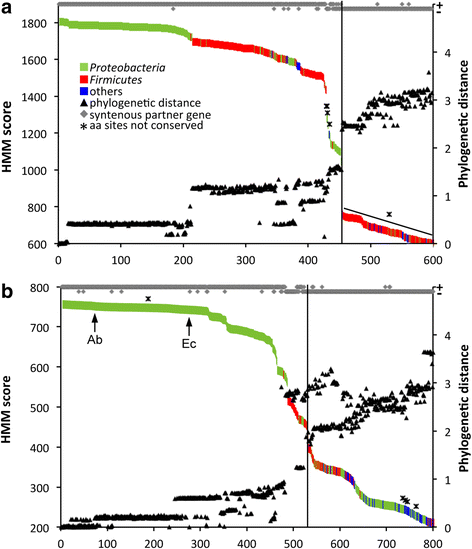
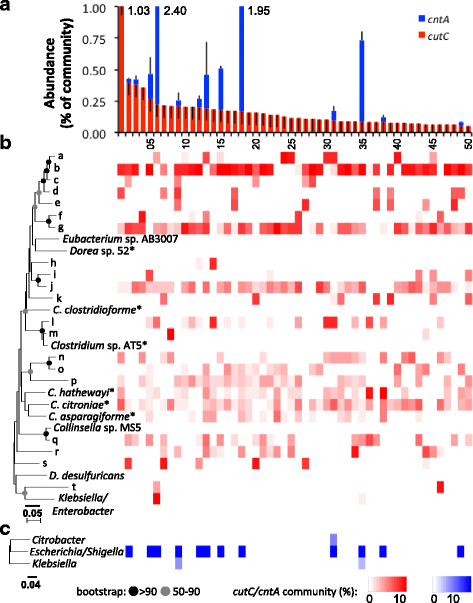
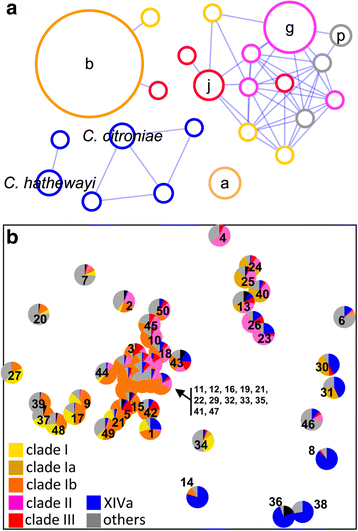
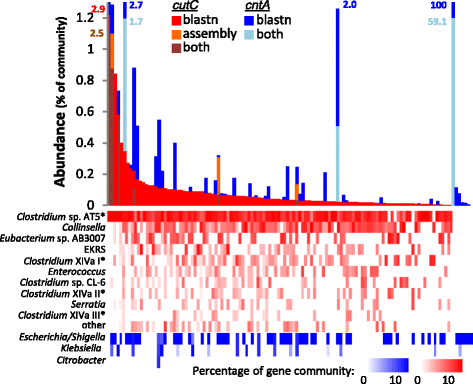
Results: In order to examine the TMA-forming potential of microbial communities, we established databases for the key genes of the main TMA-synthesis pathways, encoding choline TMA-lyase (cutC) and carnitine oxygenase (cntA), using a multi-level screening approach on 67,134 genomes revealing 1107 and 6738 candidates to exhibit cutC and cntA, respectively. Gene-targeted assays enumerating the TMA-producing community by quantitative PCR and characterizing its composition via Illumina sequencing were developed and applied on human fecal samples (n = 50) where all samples contained potential TMA producers (cutC was detected in all individuals, whereas only 26% harbored cntA) constituting, however, only a minor part of the total community (below 1% in most samples). Obtained cutC amplicons were associated with various taxa, in particular with Clostridium XIVa strains and Eubacterium sp. strain AB3007, though a bulk of sequences displayed low nucleotide identities to references (average 86% ± 7%) indicating that key human TMA producers are yet to be isolated. Co-occurrence analysis revealed specific groups governing the community structure of cutC-exhibiting taxa across samples. CntA amplicons displayed high identities (~99%) to Gammaproteobacteria-derived references, primarily from Escherichia coli. Metagenomic analysis of samples provided by the Human Microbiome Project (n = 154) confirmed the abundance patterns as well as overall taxonomic compositions obtained with our assays, though at much lower resolution, whereas 16S ribosomal RNA gene sequence analysis could not adequately uncover the TMA-producing potential. V体育安卓版.
Conclusions: In this study, we developed a diagnostic framework that enabled the quantification and comprehensive characterization of the TMA-producing potential in human fecal samples V体育ios版. The key players were identified, and together with predictions on their environmental niches using functional genomics on most closely related reference strains, we provide crucial information for the development of specific treatment strategies to restrain TMA producers and limit their proliferation. .
Keywords: Atherosclerosis; Cardiovascular disease; Functional diagnostics; Gut microbiota; Microbiome; Trimethylamine VSports最新版本. .
"V体育官网入口" Figures
"V体育ios版" References
-
- Stubbs JR, House JA. Ocque AJ, Zhang S, Johnson C, Kimber C, et al. Serum trimethylamine-N-oxide is elevated in CKD and correlates with coronary atherosclerosis burden. J Am Soc Nephrol. 2015;27:305–13. doi: 10.1681/ASN.2014111063. - DOI (VSports最新版本) - PMC - PubMed
Publication types
- "VSports" Actions
MeSH terms
- "V体育官网入口" Actions
- Actions (V体育安卓版)
- Actions (V体育2025版)
- Actions (V体育官网入口)
- Actions (V体育2025版)
- "V体育2025版" Actions
- "V体育平台登录" Actions
- "VSports注册入口" Actions
- "VSports最新版本" Actions
- "V体育官网入口" Actions
"VSports在线直播" Substances
- Actions (VSports在线直播)
- Actions (VSports在线直播)
LinkOut - more resources (V体育2025版)
Full Text Sources
Other Literature Sources
Molecular Biology Databases

