Genomic profiles of low-grade murine gliomas evolve during progression to glioblastoma
- PMID: 28398584
- PMCID: PMC5570221
- DOI: 10.1093/neuonc/nox050
VSports在线直播 - Genomic profiles of low-grade murine gliomas evolve during progression to glioblastoma
Abstract
Background: Gliomas are diverse neoplasms with multiple molecular subtypes. How tumor-initiating mutations relate to molecular subtypes as these tumors evolve during malignant progression remains unclear VSports手机版. .
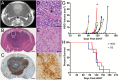
Methods: We used genetically engineered mouse models, histopathology, genetic lineage tracing, expression profiling, and copy number analyses to examine how genomic tumor diversity evolves during the course of malignant progression from low- to high-grade disease V体育安卓版. .
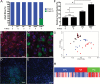
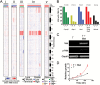
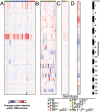
Results: Knockout of all 3 retinoblastoma (Rb) family proteins was required to initiate low-grade tumors in adult mouse astrocytes. Mutations activating mitogen-activated protein kinase signaling, specifically KrasG12D, potentiated Rb-mediated tumorigenesis. Low-grade tumors showed mutant Kras-specific transcriptome profiles but lacked copy number mutations. These tumors stochastically progressed to high-grade, in part through acquisition of copy number mutations V体育ios版. High-grade tumor transcriptomes were heterogeneous and consisted of 3 subtypes that mimicked human mesenchymal, proneural, and neural glioblastomas. Subtypes were confirmed in validation sets of high-grade mouse tumors initiated by different driver mutations as well as human patient-derived xenograft models and glioblastoma tumors. .
Conclusion: These results suggest that oncogenic driver mutations influence the genomic profiles of low-grade tumors and that these, as well as progression-acquired mutations, contribute strongly to the genomic heterogeneity across high-grade tumors VSports最新版本. .
Keywords: genetically engineered mouse; glioblastoma; glioma; progression; transcriptome. V体育平台登录.
© The Author(s) 2017. Published by Oxford University Press on behalf of the Society for Neuro-Oncology. All rights reserved. For permissions, please e-mail: journals.permissions@qiuluzeuv.cn VSports注册入口.
Figures


V体育官网 - References
-
- Miller CR, Perry A. Glioblastoma. Arch Pathol Lab Med. 2007;131(3):397–406. - PubMed
-
- Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. - "VSports" PubMed
-
- Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. - "V体育官网入口" PMC - PubMed
MeSH terms
- Actions (VSports app下载)
- VSports在线直播 - Actions
- Actions (V体育ios版)
- Actions (V体育ios版)
- "V体育官网入口" Actions
- VSports - Actions
- Actions (VSports)
- "V体育平台登录" Actions
- Actions (VSports)
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
"V体育官网入口" Medical
Molecular Biology Databases
Research Materials
Miscellaneous

