Analysis of the human monocyte-derived macrophage transcriptome and response to lipopolysaccharide provides new insights into genetic aetiology of inflammatory bowel disease (V体育2025版)
- PMID: 28263993
- PMCID: "V体育ios版" PMC5358891
- DOI: 10.1371/journal.pgen.1006641 (VSports在线直播)
Analysis of the human monocyte-derived macrophage transcriptome and response to lipopolysaccharide provides new insights into genetic aetiology of inflammatory bowel disease
Abstract
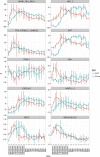
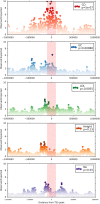
The FANTOM5 consortium utilised cap analysis of gene expression (CAGE) to provide an unprecedented insight into transcriptional regulation in human cells and tissues. In the current study, we have used CAGE-based transcriptional profiling on an extended dense time course of the response of human monocyte-derived macrophages grown in macrophage colony-stimulating factor (CSF1) to bacterial lipopolysaccharide (LPS). We propose that this system provides a model for the differentiation and adaptation of monocytes entering the intestinal lamina propria. The response to LPS is shown to be a cascade of successive waves of transient gene expression extending over at least 48 hours, with hundreds of positive and negative regulatory loops. Promoter analysis using motif activity response analysis (MARA) identified some of the transcription factors likely to be responsible for the temporal profile of transcriptional activation. Each LPS-inducible locus was associated with multiple inducible enhancers, and in each case, transient eRNA transcription at multiple sites detected by CAGE preceded the appearance of promoter-associated transcripts. LPS-inducible long non-coding RNAs were commonly associated with clusters of inducible enhancers. We used these data to re-examine the hundreds of loci associated with susceptibility to inflammatory bowel disease (IBD) in genome-wide association studies VSports手机版. Loci associated with IBD were strongly and specifically (relative to rheumatoid arthritis and unrelated traits) enriched for promoters that were regulated in monocyte differentiation or activation. Amongst previously-identified IBD susceptibility loci, the vast majority contained at least one promoter that was regulated in CSF1-dependent monocyte-macrophage transitions and/or in response to LPS. On this basis, we concluded that IBD loci are strongly-enriched for monocyte-specific genes, and identified at least 134 additional candidate genes associated with IBD susceptibility from reanalysis of published GWA studies. We propose that dysregulation of monocyte adaptation to the environment of the gastrointestinal mucosa is the key process leading to inflammatory bowel disease. .
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

V体育2025版 - References
-
- Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, et al. (2012) Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491: 119–124. 10.1038/nature11582 - DOI (VSports手机版) - PMC - PubMed
Publication types
MeSH terms
- V体育官网入口 - Actions
- VSports注册入口 - Actions
- Actions (VSports在线直播)
- VSports app下载 - Actions
- "V体育ios版" Actions
- "VSports最新版本" Actions
- Actions (VSports在线直播)
- "VSports app下载" Actions
- "V体育ios版" Actions
- VSports app下载 - Actions
- "VSports手机版" Actions
- "V体育平台登录" Actions
- Actions (VSports手机版)
- "V体育官网入口" Actions
- "V体育官网入口" Actions
Substances
- "V体育平台登录" Actions
- "VSports在线直播" Actions
- Actions (V体育安卓版)
Grants and funding (V体育平台登录)
- BB/I025328/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- VSports最新版本 - BBS/E/D/20211553/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- "VSports" BBS/E/D/20211552/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- "V体育官网入口" BB/G004013/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- MR/M019969/1/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
VSports - Other Literature Sources
Research Materials
Miscellaneous

