"V体育官网入口" Impact of Westernized Diet on Gut Microbiota in Children on Leyte Island
- PMID: 28261164
- PMCID: PMC5306386
- DOI: VSports在线直播 - 10.3389/fmicb.2017.00197
Impact of Westernized Diet on Gut Microbiota in Children on Leyte Island
Abstract
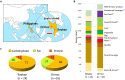
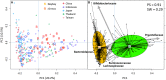
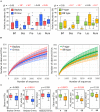
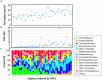
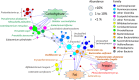
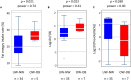
Urbanization has changed life styles of the children in some towns and cities on Leyte island in the Philippines. To evaluate the impact of modernization in dietary habits on gut microbiota, we compared fecal microbiota of 7 to 9-year-old children from rural Baybay city (n = 24) and urban Ormoc city (n = 19), and assessed the correlation between bacterial composition and diet. A dietary survey indicated that Ormoc children consumed fast food frequently and more meat and confectionary than Baybay children, suggesting modernization/westernization of dietary habits. Fat intake accounted for 27. 2% of the total energy intake in Ormoc children; this was remarkably higher than in their Baybay counterparts (18. 1%) and close to the upper limit (30%) recommended by the World Health Organization. Their fecal microbiota were analyzed by high-throughput 16S rRNA gene sequencing in conjunction with a dataset from five other Asian countries. Their microbiota were classified into two enterotype-like clusters with the other countries' children, each defined by high abundance of either Prevotellaceae (P-type) or Bacteroidaceae (BB-type), respectively. Baybay and Ormoc children mainly harbored P-type and BB-type, respectively. Redundancy analysis showed that P-type favored carbohydrates whereas BB-type preferred fats. Fat intake correlated positively with the Firmicutes-to-Bacteroidetes (F/B) ratio and negatively with the relative abundance of the family Prevotellaceae/genus Prevotella. A species-level analysis suggested that dietary fat positively correlated with an Oscillibacter species as well as a series of Bacteroides/Parabacteroides species, whereas dietary carbohydrate positively correlated with Dialister succinatiphilus known as succinate-utilizing bacteria and some succinate-producing species of family Prevotellaceae, Veillonellaceae, and Erysipelotrichaceae VSports手机版. We also found that a Succinivibrio species was overrepresented in the P-type community, suggesting the syntroph via hydrogen and succinate. Predicted metagenomics suggests that BB-type microbiota is well nourished and metabolically more active with simple sugars, amino acids, and lipids, while P-type community is more involved in digestion of complex carbohydrates. Overweight and obese children living in Ormoc, who consumed a high-fat diet, harbored microbiota with higher F/B ratio and low abundance of Prevotella. The altered gut microbiota may be a sign of a modern diet-associated obesity among children in developing areas. .
Keywords: 16S rRNA gene sequencing; Bacteroidetes; Firmicutes; Philippines; Prevotella; gut microbiota; high-fat diet; obesity. V体育安卓版.
VSports app下载 - Figures


"V体育ios版" References
-
- Backhed F., Ley R. E., Sonnenburg J. L., Peterson D. A., Gordon J. I. (2005). Host-bacterial mutualism in the human intestine. Science 307 1915–1920. 10.1126/science.1104816 - DOI (VSports) - PubMed
-
- Bäckhed F., Manchester J. K., Semenkovich C. F., Gordon J. I. (2007). Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc. Natl. Acad. Sci. U.S.A. 104 979–984. 10.1073/pnas.0605374104 - V体育官网 - DOI - PMC - PubMed
-
- Bell D. S. H. (2015). Changes seen in gut bacteria content and distribution with obesity: causation or association? Postgrad. Med. 127 863–868. 10.1080/00325481.2015.1098519 - DOI (VSports最新版本) - PubMed
LinkOut - more resources
Full Text Sources
V体育官网 - Other Literature Sources

