ZKSCAN1 gene and its related circular RNA (circZKSCAN1) both inhibit hepatocellular carcinoma cell growth, migration, and invasion but through different signaling pathways
- PMID: 28211215
- PMCID: PMC5527481
- DOI: 10.1002/1878-0261.12045
V体育ios版 - ZKSCAN1 gene and its related circular RNA (circZKSCAN1) both inhibit hepatocellular carcinoma cell growth, migration, and invasion but through different signaling pathways
Erratum in (VSports注册入口)
-
Corrigendum to: ZKSCAN1 gene and its related circular RNA (circZKSCAN1) both inhibit hepatocellular carcinoma cell growth, migration, and invasion but through different signaling pathways.Mol Oncol. 2019 Nov;13(11):2511. doi: 10.1002/1878-0261.12578. Epub 2019 Oct 7. Mol Oncol. 2019. PMID: 31670487 Free PMC article. No abstract available.
Abstract (VSports)
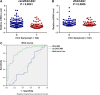
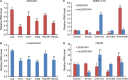
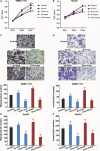
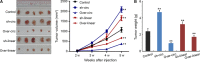
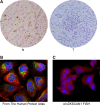
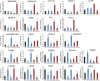
There is increasing evidence that circular RNA (circRNA) are involved in cancer development, but the regulation and function of human circRNA remain largely unknown. In this study, we demonstrated that ZKSCAN1, a zinc finger family gene, is expressed in both linear and circular (circZKSCAN1) forms of RNA in human hepatocellular carcinoma (HCC) tissues and cell lines. Here, we analyzed a cohort of 102 patients and found that expression of both ZKSCAN1mRNA and circZKSCAN1 was significantly lower (P < 0. 05) in the HCC samples compared with that in matched adjacent nontumorous tissues by reverse transcription PCR (RT-PCR). The low expression level of ZKSCAN1 was only associated with tumor size (P = 0. 032), while the cirZKSCAN1 levels varied in patients with different tumor numbers (P < 0 VSports手机版. 01), cirrhosis (P = 0. 031), vascular invasion (P = 0. 002), or microscopic vascular invasion (P = 0. 002), as well as with the tumor grade (P < 0. 001). Silencing both ZKSCAN1mRNA and circZKSCAN1 promoted cell proliferation, migration, and invasion. In contrast, overexpression of both forms of RNA repressed HCC progression in vivo and in vitro. Silencing or overexpression of both forms of RNA did not interfere with each other. RNA-seq revealed a very different molecular basis for the observed effects; ZKSCAN1mRNA mainly regulated cellular metabolism, while circZKSCAN1 mediated several cancer-related signaling pathways, suggesting a nonredundant role for ZKSCAN1mRNA and circRNA. In conclusion, our results revealed two post-translational products (ZKSCAN1mRNA and circZKSCAN1) that cooperated closely with one another to inhibit growth, migration, and invasion of HCC. cirZKSCAN1 might be a useful marker for the diagnosis of HCC. .
Keywords: HCC; RNA-seq; ZKSCAN1; circular RNA; qRT-PCR. V体育安卓版.
© 2017 The Authors. Published by FEBS Press and John Wiley & Sons Ltd V体育ios版. .
"V体育安卓版" Figures


References
-
- Audic S and Claverie JM (1997) The significance of digital gene expression profiles. Genome Res 7, 986–995. - PubMed
-
- Burd CE, Jeck WR, Liu Y, Sanoff HK, Wang Z, Sharpless NE (2010) Expression of linear and novel circular forms of an INK4/ARF‐associated non‐coding RNA correlates with atherosclerosis risk. PLoS Genet 6, e1001233. - "V体育安卓版" PMC - PubMed
-
- van Dekken H, Tilanus HW, Hop WC, Dinjens WN, Wink JC, Vissers KJ and van Marion R (2009) Array comparative genomic hybridization, expression array, and protein analysis of critical regions on chromosome arms 1q, 7q, and 8p in adenocarcinomas of the gastroesophageal junction. Cancer Genet Cytogenet 189, 37–42. - PubMed
MeSH terms
- "VSports最新版本" Actions
- V体育官网入口 - Actions
- V体育官网入口 - Actions
- "VSports手机版" Actions
- "V体育官网入口" Actions
- VSports - Actions
- "V体育安卓版" Actions
- Actions (V体育安卓版)
- "V体育平台登录" Actions
- "V体育2025版" Actions
- "VSports注册入口" Actions
- Actions (V体育ios版)
- V体育官网 - Actions
- V体育官网入口 - Actions
Substances
- Actions (VSports app下载)
- "V体育安卓版" Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

