Genetic regulatory signatures underlying islet gene expression and type 2 diabetes
- PMID: 28193859
- PMCID: "VSports最新版本" PMC5338551
- DOI: 10.1073/pnas.1621192114
Genetic regulatory signatures underlying islet gene expression and type 2 diabetes
V体育平台登录 - Abstract
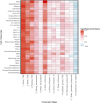
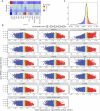
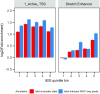
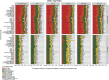
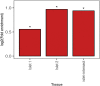
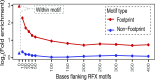
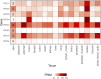
Genome-wide association studies (GWAS) have identified >100 independent SNPs that modulate the risk of type 2 diabetes (T2D) and related traits. However, the pathogenic mechanisms of most of these SNPs remain elusive. Here, we examined genomic, epigenomic, and transcriptomic profiles in human pancreatic islets to understand the links between genetic variation, chromatin landscape, and gene expression in the context of T2D VSports手机版. We first integrated genome and transcriptome variation across 112 islet samples to produce dense cis-expression quantitative trait loci (cis-eQTL) maps. Additional integration with chromatin-state maps for islets and other diverse tissue types revealed that cis-eQTLs for islet-specific genes are specifically and significantly enriched in islet stretch enhancers. High-resolution chromatin accessibility profiling using assay for transposase-accessible chromatin sequencing (ATAC-seq) in two islet samples enabled us to identify specific transcription factor (TF) footprints embedded in active regulatory elements, which are highly enriched for islet cis-eQTL. Aggregate allelic bias signatures in TF footprints enabled us de novo to reconstruct TF binding affinities genetically, which support the high-quality nature of the TF footprint predictions. Interestingly, we found that T2D GWAS loci were strikingly and specifically enriched in islet Regulatory Factor X (RFX) footprints. Remarkably, within and across independent loci, T2D risk alleles that overlap with RFX footprints uniformly disrupt the RFX motifs at high-information content positions. Together, these results suggest that common regulatory variations have shaped islet TF footprints and the transcriptome and that a confluent RFX regulatory grammar plays a significant role in the genetic component of T2D predisposition. .
Keywords: chromatin; diabetes; eQTL; epigenome; footprint V体育安卓版. .
"VSports app下载" Conflict of interest statement
The authors declare no conflict of interest.
Figures (V体育官网入口)







References
Publication types
- "V体育官网入口" Actions
"V体育官网" MeSH terms
- Actions (VSports手机版)
- "VSports在线直播" Actions
- "V体育官网" Actions
- "V体育2025版" Actions
- "V体育平台登录" Actions
- "V体育安卓版" Actions
- Actions (V体育官网入口)
- "VSports注册入口" Actions
- V体育官网入口 - Actions
- Actions (V体育官网入口)
Substances (V体育官网)
- VSports app下载 - Actions
Grants and funding
- Z01 HG000024/ImNIH/Intramural NIH HHS/United States
- R01 DK072193/DK/NIDDK NIH HHS/United States
- "V体育2025版" T32 HG000040/HG/NHGRI NIH HHS/United States
- ZIA HG000024/ImNIH/Intramural NIH HHS/United States (V体育ios版)
- U01 DK062370/DK/NIDDK NIH HHS/United States (V体育2025版)
- P30 DK020572/DK/NIDDK NIH HHS/United States
- "VSports app下载" U01 DK105561/DK/NIDDK NIH HHS/United States
- "VSports手机版" R01 DK093757/DK/NIDDK NIH HHS/United States
- "VSports最新版本" F31 HL127984/HL/NHLBI NIH HHS/United States
- R00 DK092251/DK/NIDDK NIH HHS/United States
- R00 DK099240/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
VSports最新版本 - Medical
Molecular Biology Databases
Miscellaneous

