BMP restricts stemness of intestinal Lgr5+ stem cells by directly suppressing their signature genes
- PMID: 28059064
- PMCID: PMC5227110
- DOI: 10.1038/ncomms13824
BMP restricts stemness of intestinal Lgr5+ stem cells by directly suppressing their signature genes
Abstract (VSports app下载)
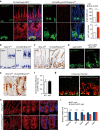
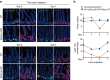
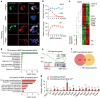
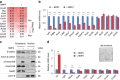
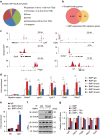
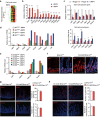
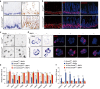
The intestinal epithelium possesses a remarkable self-renewal ability, which is mediated by actively proliferating Lgr5+ stem cells. Bone morphogenetic protein (BMP) signalling represents one major counterforce that limits the hyperproliferation of intestinal epithelium, but the exact mechanism remains elusive. Here we demonstrate that epithelial BMP signalling plays an indispensable role in restricting Lgr5+ stem cell expansion to maintain intestinal homeostasis and prevent premalignant hyperproliferation on damage VSports手机版. Mechanistically, BMP inhibits stemness of Lgr5+ stem cells through Smad-mediated transcriptional repression of a large number of stem cell signature genes, including Lgr5, and this effect is independent of Wnt/β-catenin signalling. Smad1/Smad4 recruits histone deacetylase HDAC1 to the promoters to repress transcription, and knockout of Smad4 abolishes the negative effects of BMP on stem cells. Our findings therefore demonstrate that epithelial BMP constrains the Lgr5+ stem cell self-renewal via Smad-mediated repression of stem cell signature genes to ensure proper homeostatic renewal of intestinal epithelium. .
Figures
References
-
- Barker N. et al.. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007 (2007). - V体育官网 - PubMed
-
- Vermeulen L. & Snippert H. J. Stem cell dynamics in homeostasis and cancer of the intestine. Nat. Rev. Cancer 14, 468–480 (2014). - PubMed
-
- van der Flier L. G. & Clevers H. Stem cells, self-renewal, and differentiation in the intestinal epithelium. Annu. Rev. Physiol. 71, 241–260 (2009). - PubMed
-
- Sancho E., Batlle E. & Clevers H. Signaling pathways in intestinal development and cancer. Annu. Rev. Cell Dev. Biol. 20, 695–723 (2004). - PubMed
-
- Clevers H. The intestinal crypt, a prototype stem cell compartment. Cell 154, 274–284 (2013). - PubMed
Publication types
- Actions (V体育2025版)
MeSH terms
- V体育ios版 - Actions
- Actions (V体育官网入口)
- Actions (VSports手机版)
- Actions (VSports)
- "VSports app下载" Actions
- "VSports在线直播" Actions
- "VSports注册入口" Actions
- Actions (VSports在线直播)
- V体育ios版 - Actions
- "VSports手机版" Actions
- "VSports" Actions
- Actions (VSports app下载)
- VSports注册入口 - Actions
"V体育官网" Substances
- "VSports在线直播" Actions
- Actions (VSports app下载)
LinkOut - more resources
VSports app下载 - Full Text Sources
Other Literature Sources
Medical
VSports手机版 - Molecular Biology Databases
Miscellaneous (VSports在线直播)

