CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells
- PMID: 27980086
- PMCID: PMC5394926
- DOI: 10.1126/science.aah7111
CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells (V体育官网)
V体育官网 - Abstract
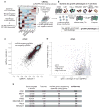
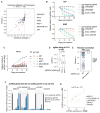
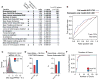
The human genome produces thousands of long noncoding RNAs (lncRNAs)-transcripts >200 nucleotides long that do not encode proteins. Although critical roles in normal biology and disease have been revealed for a subset of lncRNAs, the function of the vast majority remains untested. We developed a CRISPR interference (CRISPRi) platform targeting 16,401 lncRNA loci in seven diverse cell lines, including six transformed cell lines and human induced pluripotent stem cells (iPSCs). Large-scale screening identified 499 lncRNA loci required for robust cellular growth, of which 89% showed growth-modifying function exclusively in one cell type. We further found that lncRNA knockdown can perturb complex transcriptional networks in a cell type-specific manner. These data underscore the functional importance and cell type specificity of many lncRNAs VSports手机版. .
Copyright © 2017, American Association for the Advancement of Science. V体育安卓版.
"VSports app下载" Figures


"V体育安卓版" Comment in
-
V体育ios版 - Functional genomics: Screening for lncRNA function.Nat Rev Genet. 2017 Feb;18(2):70. doi: 10.1038/nrg.2016.168. Epub 2017 Jan 3. Nat Rev Genet. 2017. PMID: 28045101 No abstract available.
References
-
- Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. - PMC (V体育2025版) - PubMed
-
- Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. - PubMed
V体育官网 - Publication types
MeSH terms
- Actions (V体育官网)
- VSports app下载 - Actions
- Actions (VSports注册入口)
- Actions (VSports手机版)
- Actions (VSports在线直播)
- V体育ios版 - Actions
- V体育官网 - Actions
- "VSports" Actions
- Actions (VSports)
- "V体育官网入口" Actions
- "V体育官网" Actions
Substances
Grants and funding
- P01 HL089707/HL/NHLBI NIH HHS/United States
- "V体育平台登录" U01 CA168370/CA/NCI NIH HHS/United States
- T32 GM007618/GM/NIGMS NIH HHS/United States
- K99 CA204602/CA/NCI NIH HHS/United States
- P50 HG007735/HG/NHGRI NIH HHS/United States
- "VSports app下载" R01 HL130533/HL/NHLBI NIH HHS/United States
- R35 CA209919/CA/NCI NIH HHS/United States
- R00 CA204602/CA/NCI NIH HHS/United States
- T32 CA151022/CA/NCI NIH HHS/United States
- U01 HL100406/HL/NHLBI NIH HHS/United States
- F30 NS092319/NS/NINDS NIH HHS/United States
- P50 GM102706/GM/NIGMS NIH HHS/United States
- R01 DA036858/DA/NIDA NIH HHS/United States (VSports最新版本)
- "V体育官网入口" I01 BX000252/BX/BLRD VA/United States
- VSports最新版本 - R01 NS091544/NS/NINDS NIH HHS/United States
- T32 HD007470/HD/NICHD NIH HHS/United States (V体育2025版)
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials (V体育平台登录)

