TET proteins regulate the lineage specification and TCR-mediated expansion of iNKT cells
- PMID: 27869820
- PMCID: PMC5376256
- DOI: 10.1038/ni.3630
TET proteins regulate the lineage specification and TCR-mediated expansion of iNKT cells (V体育官网入口)
Abstract
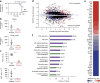
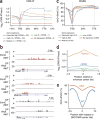
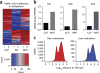
TET proteins oxidize 5-methylcytosine in DNA to 5-hydroxymethylcytosine and other oxidation products. We found that simultaneous deletion of Tet2 and Tet3 in mouse CD4+CD8+ double-positive thymocytes resulted in dysregulated development and proliferation of invariant natural killer T cells (iNKT cells) VSports手机版. Tet2-Tet3 double-knockout (DKO) iNKT cells displayed pronounced skewing toward the NKT17 lineage, with increased DNA methylation and impaired expression of genes encoding the key lineage-specifying factors T-bet and ThPOK. Transfer of purified Tet2-Tet3 DKO iNKT cells into immunocompetent recipient mice resulted in an uncontrolled expansion that was dependent on the nonclassical major histocompatibility complex (MHC) protein CD1d, which presents lipid antigens to iNKT cells. Our data indicate that TET proteins regulate iNKT cell fate by ensuring their proper development and maturation and by suppressing aberrant proliferation mediated by the T cell antigen receptor (TCR). .
Conflict of interest statement
The authors declare no competing financial interests.
Figures


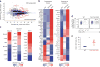

References
-
- Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol. 2007;25:297–336. - PubMed (V体育安卓版)
VSports手机版 - MeSH terms
- "V体育ios版" Actions
- "VSports最新版本" Actions
- "V体育平台登录" Actions
- VSports最新版本 - Actions
- VSports - Actions
- VSports最新版本 - Actions
- V体育平台登录 - Actions
- VSports - Actions
- Actions (VSports最新版本)
- "V体育平台登录" Actions
Substances
- Actions (V体育平台登录)
- V体育官网入口 - Actions
- Actions (VSports)
- "V体育官网" Actions
- V体育2025版 - Actions
Grants and funding
"VSports app下载" LinkOut - more resources
Full Text Sources (V体育安卓版)
"V体育平台登录" Other Literature Sources
Molecular Biology Databases
"V体育官网入口" Research Materials

