Prostaglandin E2 promotes intestinal repair through an adaptive cellular response of the epithelium (VSports app下载)
- PMID: 27797821
- PMCID: PMC5210160
- DOI: VSports在线直播 - 10.15252/embj.201694660
Prostaglandin E2 promotes intestinal repair through an adaptive cellular response of the epithelium
Abstract
Adaptive cellular responses are often required during wound repair VSports手机版. Following disruption of the intestinal epithelium, wound-associated epithelial (WAE) cells form the initial barrier over the wound. Our goal was to determine the critical factor that promotes WAE cell differentiation. Using an adaptation of our in vitro primary epithelial cell culture system, we found that prostaglandin E2 (PGE2) signaling through one of its receptors, Ptger4, was sufficient to drive a differentiation state morphologically and transcriptionally similar to in vivo WAE cells. WAE cell differentiation was a permanent state and dominant over enterocyte differentiation in plasticity experiments. WAE cell differentiation was triggered by nuclear β-catenin signaling independent of canonical Wnt signaling. Creation of WAE cells via the PGE2-Ptger4 pathway was required in vivo, as mice with loss of Ptger4 in the intestinal epithelium did not produce WAE cells and exhibited impaired wound repair. Our results demonstrate a mechanism by which WAE cells are formed by PGE2 and suggest a process of adaptive cellular reprogramming of the intestinal epithelium that occurs to ensure proper repair to injury. .
Keywords: epithelium; prostaglandin; restitution; stem cell; wound repair. V体育安卓版.
© 2016 The Authors.
Figures (VSports)
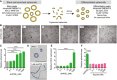
- A
Schematic of spheroid differentiation method. L‐WRN CM, conditioned medium from an L cell line engineered to secrete Wnt3a, R‐spondin 3, and noggin. Y‐27632, ROCK inhibitor. EGF, epidermal growth factor V体育平台登录.
- B–F
Mouse jejunal spheroids were cultured in differentiation medium containing 0–1,000 nM dmPGE2 or iloprost. (B) Representative bright‐field images of spheroids treated with dmPGE2. Scale bars, 200 μm. (C) Quantification of average spheroid area ± s. e VSports注册入口. m. relative to spheroids treated with 0 nM dmPGE2 (average area was 3,761 μm2 for 0 nM group; n = 4 images with a minimum of 50 spheroids counted from two independent experiments). (D) Representative histological sections of spheroids treated with 1 μM dmPGE2 or an equivalent volume of DMSO and stained with hematoxylin. Scale bars, 100 μm. (E) Quantification of the average expression ± s. e. m. of Cldn4 mRNA relative to the 0 nM treatment group (n = 3 independent experiments). (F) Quantification of average spheroid area ± s. e. m. relative to spheroids treated with 0 nM iloprost (n = 4 images with a minimum of 50 spheroids counted from two independent experiments). ***P < 0. 001, ****P < 0. 0001 by one‐way ANOVA and Dunnett's post‐test.

- A
Quantification of the average expression ± s. e. m. of Ptger1, Ptger2, Ptger3, Ptger4, and Ptgir mRNAs in whole‐thickness mouse lung, ileum, or colon tissues or in jejunal spheroids cultured in stem cell (Stem), WAE (dmPGE2), or enterocyte (EP4i) medium (n = 3 independent experiments). Data are presented as fold change compared to lung tissue. n. d. , not detected VSports在线直播.
- B–D
Mouse jejunal spheroids were cultured in differentiation medium with DMSO only or with 1 μM dmPGE2 and pharmacological inhibitors of EP1 (EP1i, SC 51322), EP2 (EP2i, PF 04418948), EP3 (EP3i, L‐798,106), or EP4 (EP4i, L‐161,982) at a concentration of 10 μM or an equivalent volume of DMSO vehicle. **P < 0.01, ***P < 0.001, ****P < 0.0001 compared to the DMSO only group by one‐way ANOVA and Dunnett's post‐test. (B) Representative bright‐field images. Scale bars, 200 μm. (C) Quantification of average spheroid area ± s.e.m. relative to spheroids treated with DMSO alone (n = 4 images with a minimum of 50 spheroids counted from two independent experiments). (D) Quantification of the average expression ± s.e.m. of Cldn4 mRNA relative to stem cell spheroids (n = 3 independent experiments).
- E, F
Tat‐Cre mediated recombination of Ptger4 flox/flox (fl/fl) jejunal spheroids to generate Ptger4 flox/Δ (fl/Δ) and Ptger4 Δ/Δ (Δ/Δ) spheroid lines. (E) Schematic and representative PCR genotyping results. M, marker lane. bp, base pairs. (F) Quantification of the average expression ± s.e.m. of Ptger4 mRNA in spheroids cultured in stem cell media relative to fl/fl genotype (n = 3 independent experiments). ***P < 0.001 by two‐way ANOVA and Dunnett's multiple comparisons post‐test with the EP4i‐treated fl/fl group set as the control.
- G–I
Ptger4 flox/flox, Ptger4 flox/Δ, and Ptger4 Δ/Δ spheroids were cultured in differentiation medium with 1 μM dmPGE2 or 10 μM EP4i. **P < 0.01, ****P < 0.0001 by one‐way ANOVA and Tukey's post‐test. (G) Representative bright‐field images. Scale bars, 200 μm. (H) Quantification of average spheroid area ± s.e.m. (n = 4 images with a minimum of 50 spheroids counted per group from two independent experiments) and (I) average expression ± s.e.m. of Cldn4 mRNA (n = 3 independent experiments) relative to EP4i‐treated Ptger4 flox/flox spheroids.
- J–L
Human ileal spheroids were cultured in differentiation medium with 1 μM dmPGE2 or 10 μM EP4i. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 by one‐way ANOVA and Tukey's post‐test. (J) Representative bright‐field images. Scale bars, 200 μm. (K) Quantification of average spheroid area ± s.e.m. (n = 4 images with a minimum of 50 spheroids counted per group from three independent donor lines examined over 3 passages each) and (L) the average expression ± s.e.m. of CLDN4 mRNA (n = 3 independent donor lines) relative to stem spheroids.
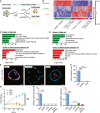
- A
Schematic of method used to generate stem cell, enterocyte, and WAE cell groups.
- B
Heatmap showing unsupervised hierarchical clustering of genes that were significantly (FDR‐adjusted P < 0.05) and differentially expressed between the three groups (n = 4 samples per treatment from two independent experiments). Each column is one sample and each row is one gene. Red and blue indicate genes with enriched or de‐enriched mRNA expression in a particular group, respectively. The dendrogram for the gene clustering is colored to highlight major gene clusters 1–6. The number of probes in each cluster is in parentheses.
- C, D
Graphs showing the top five most significant pathways (C) and gene ontology cellular component terms (D) associated with Cluster 1 and Cluster 4.
- E, F
Spheroids were cultured as indicated for 24 h followed by a 1‐h pulse with EdU to mark the cells undergoing DNA synthesis. (E) Representative images of EdU staining (red). Nuclei are visualized with bisbenzimide (blue). Scale bars, 20 μm. (F) Quantification of EdU‐positive nuclei ± s.e.m. as a percent of the total nuclei (n = minimum of 15 spheroids counted per sample from two independent experiments). ****P < 0.0001 by one‐way ANOVA and Tukey's post‐test.
- G
Graph of the fold change in background‐subtracted luminescence ± s.e.m. (relative to 0 h measurement) of Cdc25A‐CBRLuc spheroids (n = 3 independent experiments with four technical replicates). ****P < 0.0001 for dmPGE2‐ and EP4i‐treated spheroids compared to stem cells by repeated measures two‐way ANOVA (variable = treatment). P < 0.001 at the 16, 20, and 24 h time points by Dunnett's post‐test comparing dmPGE2‐treated and EP4i‐treated spheroids to the stem cell control.
- H
Quantification of the average expression ± s.e.m. of Lgr5 and Mki67 mRNAs in jejunal spheroids cultured in stem cell (Stem) or in differentiation medium with the indicated supplements relative to the stem cell group (n = 3 independent experiments). **P < 0.01, ****P < 0.0001 by one‐way ANOVA and Dunnett's post‐test. n.d., not detected.
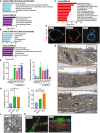
- A, B
Graphs showing the top five most significant pathways (A) and gene ontology cellular component terms (B) associated with Cluster 5 and Cluster 6.
- C
Graph of the top twelve significantly enriched pathways in in vivo colonic WAE cells.
- D
Representative images of spheroids stained for Cldn4 (red). Nuclei are visualized with bisbenzimide (blue) (n = 2 independent experiments). Scale bars, 20 μm.
- E, F
Jejunal spheroids were cultured as in Fig 2B–D. Quantification of the average expression ± s.e.m. of Dpcr1 (E) and Cd55b mRNAs relative to DMSO group (n = 3 independent experiments). *P < 0.05, ****P < 0.0001 by one‐way ANOVA and Dunnett's post‐test.
- G–J
Representative transmission electron microscopy (TEM) images of stem and WAE spheroids and an in vivo WAE cell from a biopsy‐injured mouse colon. (G) The basal plasma membranes are outlined in orange solid lines, lateral plasma membranes are indicated with orange arrowheads, and nuclei are outlined with wide yellow dashed lines. Insets show a magnified view of the apical cell surface. Quantification of cytoplasmic:nuclear ratio (H) and microvillar length (I) ± s.e.m. from the TEM images (n = minimum of five images per group). *P < 0.05, **P < 0.01, ****P < 0.0001 by one‐way ANOVA and Tukey's post‐test. (J) Higher power image of the cytoplasm of a WAE spheroid cell. Single mitochondria are outlined with a narrow blue dashed line; vacuole structures are indicated with red asterisks. Scale bars, 1 μm.
- K
Representative image of a spheroid stained for β‐catenin (green) and F‐actin (red). Nuclei are visualized with bisbenzimide (blue). A similarly stained section of a small intestinal ulcer is shown for comparison. Arrowheads indicate the apical cell membrane. Scale bars, 50 μm.
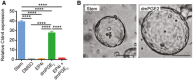
Quantification of the average expression ± s.e.m. of Cldn4 mRNA in mouse jejunal spheroids cultured in stem cell or in differentiation medium with the indicated supplements relative to the DMSO group (n = 3 independent experiments). ****P < 0.0001 by one‐way ANOVA and Tukey's post‐test.
Representative bright‐field images of stem or dmPGE2‐treated spheroids. Stem spheroids have a smooth appearance, whereas dmPGE2‐treated spheroids have a dimpled “golf ball‐like” appearance. Scale bars, 200 μm.
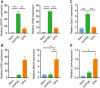
- A, B
Human ileal spheroids were cultured in stem cell medium or in differentiation medium containing dmPGE2 or EP4i. Quantification of the average mRNA expression ± s.e.m. of the WAE cell markers DPCR1 and CD55 (A) and the enterocyte markers ACE2 and MAOA (B) relative to the stem group (n = 3 independent human lines). **P < 0.01, ***P < 0.001, ****P < 0.0001 by one‐way ANOVA and Tukey's post‐test.
- C, D
Mouse colon spheroids were cultured as indicated. Quantification of the average mRNA expression ± s.e.m. of the WAE marker Dpcr1 (C) and the colonocyte marker Car4 (D) relative to stem group (n = 3 independent experiments). *P < 0.05, **P < 0.01 as determined by one‐way ANOVA and Tukey's post‐test.
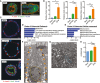
- A
Representative image of a mouse spheroid stained for β‐catenin (green) and F‐actin (red). Nuclei are visualized with bisbenzimide (blue). A similarly stained section of a mouse villus is shown for comparison. Arrowheads indicate the apical cell membrane. Scale bars, 50 μm.
- B, C
Mouse jejunal spheroids were cultured as indicated. (B) Quantification of the average expression ± s.e.m. of Fabp1, Ace2, and Maoa mRNAs (n = 3 independent experiments). *P < 0.05, **P < 0.01 by one‐way ANOVA and Tukey's post‐test. (C) Representative images of spheroids stained for Ace2 (green) and β‐catenin (red). Nuclei are visualized with bisbenzimide (blue). Scale bars, 20 μm.
- D, E
Graphs showing the top five most significant pathways (D) and gene ontology cellular component terms (E) associated with Cluster 3.
- F, G
Representative TEM images of EP4i‐treated spheroids. (F) The basal plasma membranes are outlined in orange solid lines, lateral plasma membranes are indicated with orange arrowheads, and nuclei are outlined with wide yellow dashed lines. Inset shows a magnified view of the apical cell surface. (G) Higher power image of the cytoplasm. Single mitochondria are outlined with a narrow blue dashed line. Scale bars, 1 μm.
- H
Graph of spheroid oxygen consumption rate (OCR) to extracellular acidification rate (ECAR) ratio expressed as mean ± s.e.m. (n = 3 independent experiments). **P < 0.01 by one‐way ANOVA and Tukey's post‐test.
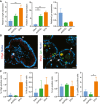
- A–C
Mouse jejunal spheroids were cultured in stem cell medium or in differentiation medium containing dmPGE2 or EP4i followed by assessment of intestinal secretory lineages. *P < 0.05, **P < 0.01 by one‐way ANOVA and Tukey's post‐test. (A) Quantification of the average mRNA expression ± s.e.m. of the endocrine marker chromogranin A (ChgA), the goblet cell marker mucin 2 (Muc2), and the Paneth cell marker lysozyme (Lyz) relative to the stem group (n = 3 independent experiments). (B) Representative fluorescent images of histological sections of spheroids stained with ChgA (red; arrowheads) or co‐stained for Muc2 (red; arrowheads) and Lyz (green; arrows) as indicated. Asterisks indicate cells that co‐expressed Muc2 and Lyz. Nuclei are visualized with bisbenzimide (blue). Scale bars, 20 μm. (C) Quantification of the cells expressing the indicated protein markers as a percentage ± s.e.m. of the total nuclei counted per group (n = minimum of three images counted from each treatment group; two independent experiments).
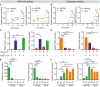
- A–D
Quantification of the average expression ± s.e.m. of Dpcr1 and Cd55b mRNAs (A, C) or Fabp1 and Ace2 mRNAs (B, D) in mouse jejunal spheroids cultured in differentiation medium containing EP4i or dmPGE2. (A, B) Gene expression analyzed 2, 6, 12, or 24 h after the start of treatment (n = 3 independent experiments). ***P < 0.001, ****P < 0.0001 comparing the two treatment groups and † P < 0.05, ††† P < 0.001, †††† P < 0.0001 compared to the 2‐h time point of the same medium by two‐way ANOVA and Sidak's multiple comparisons test. (C, D) Gene expression was analyzed after culturing spheroids in EP4i (E) or dmPGE2 (P) for the first 12 h followed by washout and re‐feeding with EP4i or dmPGE2 for the second 12 h as shown (n = 3 independent experiments). *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 by one‐way ANOVA and Dunnett's post‐test.
- E, F
Gene expression was analyzed in Ptger4 flox/flox, Ptger4 flox/Δ, and Ptger4 Δ/Δ spheroids and expressed as fold change relative to EP4i‐treated Ptger4 flox/flox (n = 3 independent experiments). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 by two‐way ANOVA and Tukey's post‐test comparing the genotypes within a treatment group. † P < 0.05, ††† P < 0.001, †††† P < 0.0001 by two‐way ANOVA and Sidak's post‐test comparing the effects of the treatments within a genotype.
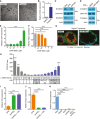
- A, B
Mouse jejunal spheroids were cultured in differentiation medium containing 10 μM of forskolin or an equivalent volume of DMSO. (A) Representative bright‐field images. Scale bars, 200 μm. (B) Quantification of the average expression ± s.e.m. of Cldn4 mRNA relative to the DMSO treatment group (n = 3 independent experiments). *P < 0.05 compared to DMSO group by unpaired t‐test.
- C, D
Representative immunoblots for β‐catenin detected in nuclear (Nuc) and cytoplasmic (Cyt) protein lysates. Lamin A/C and actin were used as loading controls for the nuclear and cytoplasmic fractions, respectively (n = 3 independent experiments). (C) Spheroids were cultured in stem, enterocyte (EP4i), or WAE (dmPGE2) media. (D) Spheroids were cultured in enterocyte medium with DMSO or 10 μM CHIR 99021.
- E, F
Spheroids were cultured in enterocyte medium with the indicated concentrations of CHIR 99021. Quantification of the average expression ± s.e.m. of Cldn4 (E) and Fabp1 (F) mRNAs shown as fold change relative to 0 μM group (n = 3 independent experiments). **P < 0.01, ****P < 0.0001 as determined by one‐way ANOVA and Dunnett's post‐test.
- G
Representative images of Ptger4 Δ/Δ spheroids cultured in differentiation medium with DMSO or 10 μM CHIR 99021 and stained for β‐catenin (green) and F‐actin (red). Nuclei are visualized with bisbenzimide (blue). Arrowheads indicate the apical cell membrane. Scale bars, 20 μm.
- H
Graph of TOPFlash (TOP) to FOPFlash (FOP) luciferase reporter ratios ± s.e.m. (n = 3 independent experiments) in transfected 293FT cells treated with conditioned mediums collected from L‐WRN cells and diluted as indicated. L‐WRN CM had been produced in the presence or absence of the porcupine inhibitor C59 (10‐fold dilutions, 10 μM to 1 pM). *P < 0.05, ***P < 0.001, ****P < 0.0001 by one‐way ANOVA and Dunnett's post‐test compared to 0% L‐WRN CM group.
- I, J
Spheroids were cultured in enterocyte media, WAE media, or WAE media containing 100 pM C59. Quantification of the average expression ± s.e.m. of Cldn4 (I) and Fabp1 (J) mRNAs shown as fold change relative to EP4i group (n = 3 independent experiments). ***P < 0.001, ****P < 0.0001 as determined by one‐way ANOVA and Tukey's post‐test.
- K
Quantification of the average expression ± s.e.m. of Axin2 mRNA in spheroids cultured in stem cell or in differentiation medium with the indicated supplements relative to stem (n = 3 independent experiments). ****P < 0.0001 compared to stem cell group as determined by one‐way ANOVA and Dunnett's post‐test.


- A
Representative whole‐mount images of wounds from Ptger4 flox/flox (Ptger4 fl/fl) and Vil Cre Ptger4 flox/flox (Vil Cre Ptger4 fl/fl) mice 4 days post‐biopsy injury. Scale bars, 750 μm. Wounds are outlined with a black dashed line. Asterisk indicates fibrin clot.
- B, C
Quantification of percent healing (1 – [day 4 wound area/original wound area] × 100) (B) and fibrin clot areas (C) ± s.e.m. (n = 7–8 mice per genotype with 3–4 wounds each, three independent experiments). *P = 0.0270, **P = 0.0057 by two‐tailed unpaired t‐test.
- D, E
Representative images of serial wound tissue sections from the indicated genotypes. (D) Fluorescent images of sections stained for Claudin‐4 (red). Nuclei are visualized with bisbenzimide (blue). Epithelial crypts adjacent to the wound bed are indicated by dashed yellow lines. Layer of adherent WAE cells indicated by white arrowheads. Non‐adherent residual WAE cells indicated by orange arrows. (E) Grayscale images of hematoxylin‐ and eosin‐stained sections. Insets of the boxed regions show a higher magnification of the region typically containing WAE cells (red arrowheads, dashed line indicates border with wound bed). Orange arrow indicates region of non‐adherent residual WAE cells. Scale bars, 100 μm.
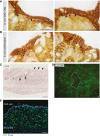
- A, B
Representative high power, bright‐field images of control mice wound tissue sections immunostained for β‐catenin (brown). Scale bars, 20 μm. (A) WAE cells covering wound bed. (B) Surface colonocytes from an uninjured area. Examples of cells with accumulation of nuclear β‐catenin are indicated by arrows, whereas examples of cells with strong membrane staining but low nuclear β‐catenin staining are indicated with arrowheads.
- C
Representative in situ hybridization result for Axin2 mRNA on day 6 post‐biopsy showing accumulation of signal at the crypt bases where stem cells reside (arrows), but no signal in WAE cells (arrowheads). Scale bar, 50 μm.
- D
Representative whole‐mount fluorescent image of colonic wound on day 6 post‐biopsy. Endogenous GFP expression is driven by the Lgr5 promoter (this genetic mouse model has mosaic GFP expression). The wounded area is outlined in a dashed white line. Scale bar, 50 μm.
- E
Representative image of control mouse wound tissue section on day 4 post‐biopsy stained for Ki67 (green). Nuclei are visualized with bisbenzimide (blue). The white dashed line separates the non‐proliferative WAE cells from the highly proliferative cells of the wound bed. Scale bar, 50 μm.
VSports最新版本 - Comment in
-
The Wae to repair: prostaglandin E2 (PGE2) triggers intestinal wound repair.EMBO J. 2017 Jan 4;36(1):3-4. doi: 10.15252/embj.201695973. Epub 2016 Dec 8. EMBO J. 2017. PMID: 27932445 Free PMC article.
References
-
- Abramovitz M, Adam M, Boie Y, Carriere M, Denis D, Godbout C, Lamontagne S, Rochette C, Sawyer N, Tremblay NM, Belley M, Gallant M, Dufresne C, Gareau Y, Ruel R, Juteau H, Labelle M, Ouimet N, Metters KM (2000) The utilization of recombinant prostanoid receptors to determine the affinities and selectivities of prostaglandins and related analogs. Biochim Biophys Acta 1483: 285–293 - PubMed
-
- Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, Clevers H (2007) Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449: 1003–1007 - PubMed
-
- Becker C, Fantini MC, Wirtz S, Nikolaev A, Kiesslich R, Lehr HA, Galle PR, Neurath MF (2005) In vivo imaging of colitis and colon cancer development in mice using high resolution chromoendoscopy. Gut 54: 950–954 - PMC (V体育官网入口) - PubMed
-
- Bielefeld KA, Amini‐Nik S, Whetstone H, Poon R, Youn A, Wang J, Alman BA (2011) Fibronectin and beta‐catenin act in a regulatory loop in dermal fibroblasts to modulate cutaneous healing. J Biol Chem 286: 27687–27697 - PMC (VSports) - PubMed
Publication types
MeSH terms
- "VSports最新版本" Actions
- Actions (V体育ios版)
- V体育ios版 - Actions
- "VSports注册入口" Actions
Substances
"V体育官网" Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

