Determining the Mitochondrial Methyl Proteome in Saccharomyces cerevisiae using Heavy Methyl SILAC
- PMID: 27696855
- PMCID: PMC5148652 (V体育平台登录)
- DOI: 10.1021/acs.jproteome.6b00521
VSports最新版本 - Determining the Mitochondrial Methyl Proteome in Saccharomyces cerevisiae using Heavy Methyl SILAC
Abstract
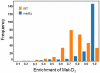
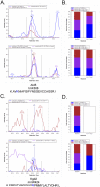
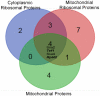
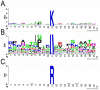
Methylation is a common and abundant post-translational modification. High-throughput proteomic investigations have reported many methylation sites from complex mixtures of proteins. The lack of consistency between parallel studies, resulting from both false positives and missed identifications, suggests problems with both over-reporting and under-reporting methylation sites. However, isotope labeling can be used effectively to address the issue of false-positives, and fractionation of proteins can increase the probability of identifying methylation sites in lower abundance. Here we have adapted heavy methyl SILAC to analyze fractions of the budding yeast Saccharomyces cerevisiae under respiratory conditions to allow for the production of mitochondria, an organelle whose proteins are often overlooked in larger methyl proteome studies. We have found 12 methylation sites on 11 mitochondrial proteins as well as an additional 14 methylation sites on 9 proteins that are nonmitochondrial. Of these methylation sites, 20 sites have not been previously reported. This study represents the first characterization of the yeast mitochondrial methyl proteome and the second proteomic investigation of global mitochondrial methylation to date in any organism VSports手机版. .
Keywords: MudPIT; heavy methyl SILAC; methyl proteome; methylation; mitochondria; protein arginine methylation; protein lysine methylation; yeast. V体育安卓版.
Figures



VSports - References
-
- Walsh C. Posttranslational Modification of Proteins: Expanding Nature’s Inventory. Roberts and Company Publishers; 2006. Protein Methylation; pp. 121–150.
-
- Biggar KK, Li SS-C. Non-histone protein methylation as a regulator of cellular signalling and function. Nat. Rev. Mol. Cell Biol. 2015;16(1):5–17. - PubMed
MeSH terms
- V体育平台登录 - Actions
- Actions (V体育安卓版)
- Actions (VSports在线直播)
- VSports在线直播 - Actions
Substances
- Actions (V体育官网)
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases (V体育安卓版)

