Neddylation E2 UBE2F Promotes the Survival of Lung Cancer Cells by Activating CRL5 to Degrade NOXA via the K11 Linkage
- PMID: 27591266
- PMCID: PMC5315595
- DOI: 10.1158/1078-0432.CCR-16-1585
Neddylation E2 UBE2F Promotes the Survival of Lung Cancer Cells by Activating CRL5 to Degrade NOXA via the K11 Linkage (VSports app下载)
Abstract
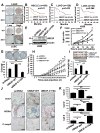
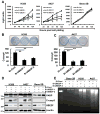
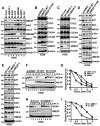
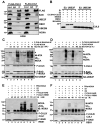
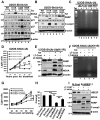
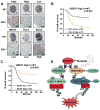
Purpose: Recent studies have shown that the process of protein neddylation was abnormally activated in several human cancers. However, it is unknown whether and how UBE2F, a less characterized neddylation E2, regulates lung cancer cell survival, and whether and how NOXA, a proapoptotic protein, is ubiquitylated and degraded by which E3 and via which ubiquitin linkage. Experimental Design: Methods of immunohistochemistry and immunoblotting were utilized to examine UBE2F protein expression. The biological functions of UBE2F were evaluated by in vitro cell culture and in vivo xenograft models. The in vivo complex formation among UBE2F-SAG-CUL5-NOXA was measured by a pulldown assay. Polyubiquitylation of NOXA was evaluated by in vivo and in vitro ubiquitylation assays. Results: UBE2F is overexpressed in non-small cell lung cancer (NSCLC) and predicts poor patient survival. While UBE2F overexpression promotes lung cancer growth both in vitro and in vivo, UBE2F knockdown selectively inhibits tumor growth. By promoting CUL5 neddylation, UBE2F/SAG/CUL5 tri-complex activates CRL5 (Cullin-RING-ligase-5) to ubiquitylate NOXA via a novel K11, but not K48, linkage for targeted proteasomal degradation VSports手机版. CRL5 inactivation or forced expression of K11R ubiquitin mutant caused NOXA accumulation to induce apoptosis, which is rescued by NOXA knockdown. Notably, NOXA knockdown rescues the UBE2F silencing effect, indicating a causal role of NOXA in this process. In lung cancer tissues, high levels of UBE2F and CUL5 correlate with a low level of NOXA and poor patient survival. Conclusions: By ubiquitylating and degrading NOXA through activating CRL5, UBE2F selectively promotes lung cancer cell survival and could, therefore, serve as a novel cancer target. Clin Cancer Res; 23(4); 1104-16. ©2016 AACR. .
©2016 American Association for Cancer Research. V体育安卓版.
Conflict of interest statement
Figures
V体育ios版 - References
-
- Kamitani T, Kito K, Nguyen HP, Yeh ET. Characterization of NEDD8, a developmentally down-regulated ubiquitin-like protein. The Journal of biological chemistry. 1997;272:28557–62. - PubMed
-
- Watson IR, Irwin MS, Ohh M. NEDD8 pathways in cancer, Sine Quibus Non. Cancer cell. 2011;19:168–76. - PubMed
-
- Zhao Y, Morgan MA, Sun Y. Targeting Neddylation Pathways to Inactivate Cullin-RING Ligases for Anticancer Therapy. Antioxidants & redox signaling. 2014;21:2383–400. - "VSports app下载" PMC - PubMed
-
- Sarkaria I, Oc P, Talbot SG, Reddy PG, Ngai I, Maghami E, et al. Squamous cell carcinoma related oncogene/DCUN1D1 is highly conserved and activated by amplification in squamous cell carcinomas. Cancer research. 2006;66:9437–44. - PubMed
-
- Singh B, Stoffel A, Gogineni S, Poluri A, Pfister DG, Shaha AR, et al. Amplification of the 3q26.3 locus is associated with progression to invasive cancer and is a negative prognostic factor in head and neck squamous cell carcinomas. The American journal of pathology. 2002;161:365–71. - PMC - PubMed
MeSH terms
- Actions (VSports注册入口)
- "V体育平台登录" Actions
- VSports注册入口 - Actions
- "VSports手机版" Actions
- "V体育ios版" Actions
- "V体育2025版" Actions
- Actions (V体育ios版)
- Actions (VSports手机版)
- Actions (VSports在线直播)
- Actions (VSports注册入口)
- V体育官网 - Actions
Substances (V体育官网)
- "VSports最新版本" Actions
"VSports" Grants and funding
"VSports在线直播" LinkOut - more resources
Full Text Sources
Other Literature Sources (VSports手机版)
"V体育官网" Medical
Molecular Biology Databases

