CD19 CAR immune pressure induces B-precursor acute lymphoblastic leukaemia lineage switch exposing inherent leukaemic plasticity
- PMID: 27460500
- PMCID: PMC4974466
- DOI: 10.1038/ncomms12320
"VSports app下载" CD19 CAR immune pressure induces B-precursor acute lymphoblastic leukaemia lineage switch exposing inherent leukaemic plasticity
Abstract
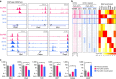
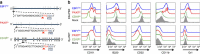
Adoptive immunotherapy using chimeric antigen receptor (CAR) expressing T cells targeting the CD19 B lineage receptor has demonstrated marked success in relapsed pre-B-cell acute lymphoblastic leukaemia (ALL). Persisting CAR-T cells generate sustained pressure against CD19 that may drive unique mechanisms of resistance. Pre-B ALL originates from a committed pre-B cell or an earlier progenitor, with potential to reprogram into other hematopoietic lineages. Here we report changes in lineage markers including myeloid conversion in patients following CD19 CAR therapy. Using murine ALL models we study the long-term effects of CD19 CAR-T cells and demonstrate partial or complete lineage switch as a consistent mechanism of CAR resistance depending on the underlying genetic oncogenic driver. Deletion of Pax5 or Ebf1 recapitulates lineage reprogramming occurring during CD19 CAR pressure. Our findings establish lineage switch as a mechanism of CAR resistance exposing inherent plasticity in genetic subtypes of pre-B-cell ALL. VSports手机版.
"VSports app下载" Figures



References
-
- Vardiman J. W. et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood 114, 937–951 (2009). - PubMed
-
- Inaba H., Greaves M. & Mullighan C. G. Acute lymphoblastic leukaemia. Lancet 381, 1943–1955 (2013). - "V体育安卓版" PMC - PubMed
-
- Meyers S. C. & Levine R. L. Translational implications of somatic genomics in acute myeloid leukaemia. Lancet Oncol. 15, e382–e394 (2014). - PubMed
-
- Greaves M. F. & Wiemels J. Origins of chromosome translocations in childhood leukaemia. Nat. Rev. Cancer 3, 639–649 (2003). - PubMed
Publication types
- "V体育安卓版" Actions
MeSH terms (V体育官网入口)
- V体育ios版 - Actions
- VSports在线直播 - Actions
- "VSports在线直播" Actions
- VSports手机版 - Actions
- "V体育安卓版" Actions
V体育官网 - Substances
- Actions (VSports app下载)
LinkOut - more resources (V体育官网)
Full Text Sources (VSports手机版)
Other Literature Sources
Molecular Biology Databases
Research Materials (VSports在线直播)

