Mechanisms of TGFβ-Induced Epithelial-Mesenchymal Transition
- PMID: 27367735
- PMCID: PMC4961994
- DOI: V体育官网 - 10.3390/jcm5070063
"V体育官网" Mechanisms of TGFβ-Induced Epithelial-Mesenchymal Transition
Abstract
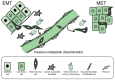
Transitory phenotypic changes such as the epithelial-mesenchymal transition (EMT) help embryonic cells to generate migratory descendants that populate new sites and establish the distinct tissues in the developing embryo. The mesenchymal descendants of diverse epithelia also participate in the wound healing response of adult tissues, and facilitate the progression of cancer. EMT can be induced by several extracellular cues in the microenvironment of a given epithelial tissue. One such cue, transforming growth factor β (TGFβ), prominently induces EMT via a group of specific transcription factors. The potency of TGFβ is partly based on its ability to perform two parallel molecular functions, i. e. to induce the expression of growth factors, cytokines and chemokines, which sequentially and in a complementary manner help to establish and maintain the EMT, and to mediate signaling crosstalk with other developmental signaling pathways, thus promoting changes in cell differentiation. The molecules that are activated by TGFβ signaling or act as cooperating partners of this pathway are impossible to exhaust within a single coherent and contemporary report. Here, we present selected examples to illustrate the key principles of the circuits that control EMT under the influence of TGFβ. VSports手机版.
Keywords: epithelial-mesenchymal transition; signal transduction; transcription factor; transforming growth factor β; tumor invasiveness V体育安卓版. .
Figures


"VSports注册入口" References
-
- Kalluri R., Weinberg R.A. The basics of epithelial-mesenchymal transition. J. Clin. Investig. 2009;119:1420–1428. doi: 10.1172/JCI39104. - VSports手机版 - DOI - PMC - PubMed
Publication types
VSports app下载 - LinkOut - more resources
Full Text Sources (V体育2025版)
Other Literature Sources

