"VSports注册入口" TGF-β Tumor Suppression through a Lethal EMT
- PMID: 26898331
- PMCID: PMC4801341
- DOI: 10.1016/j.cell.2016.01.009
"V体育平台登录" TGF-β Tumor Suppression through a Lethal EMT
Abstract
TGF-β signaling can be pro-tumorigenic or tumor suppressive VSports手机版. We investigated this duality in pancreatic ductal adenocarcinoma (PDA), which, with other gastrointestinal cancers, exhibits frequent inactivation of the TGF-β mediator Smad4. We show that TGF-β induces an epithelial-mesenchymal transition (EMT), generally considered a pro-tumorigenic event. However, in TGF-β-sensitive PDA cells, EMT becomes lethal by converting TGF-β-induced Sox4 from an enforcer of tumorigenesis into a promoter of apoptosis. This is the result of an EMT-linked remodeling of the cellular transcription factor landscape, including the repression of the gastrointestinal lineage-master regulator Klf5. Klf5 cooperates with Sox4 in oncogenesis and prevents Sox4-induced apoptosis. Smad4 is required for EMT but dispensable for Sox4 induction by TGF-β. TGF-β-induced Sox4 is thus geared to bolster progenitor identity, whereas simultaneous Smad4-dependent EMT strips Sox4 of an essential partner in oncogenesis. Our work demonstrates that TGF-β tumor suppression functions through an EMT-mediated disruption of a lineage-specific transcriptional network. .
Copyright © 2016 Elsevier Inc. All rights reserved V体育安卓版. .
Conflict of interest statement
The authors declare to have no financial interests in connection with this work.
Figures
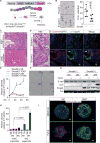
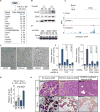
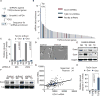



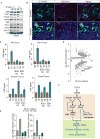
Comment in
-
EMT: Matter of Life or Death?Cell. 2016 Feb 25;164(5):840-2. doi: 10.1016/j.cell.2016.02.024. Cell. 2016. PMID: 26919422
-
V体育安卓版 - Cancer biology: TGFβ and EMT as double agents.Nat Rev Mol Cell Biol. 2016 Apr;17(4):202-3. doi: 10.1038/nrm.2016.26. Epub 2016 Mar 2. Nat Rev Mol Cell Biol. 2016. PMID: 26931318 No abstract available.
"VSports手机版" References
-
- Bialkowska AB, Liu Y, Nandan MO, Yang VW. A colon cancer-derived mutant of Kruppel-like factor 5 (KLF5) is resistant to degradation by glycogen synthase kinase 3beta (GSK3beta) and the E3 ubiquitin ligase F-box and WD repeat domain-containing 7alpha (FBW7alpha) The Journal of biological chemistry. 2014;289:5997–6005. - PMC (VSports注册入口) - PubMed
Publication types
- Actions (V体育官网)
MeSH terms
- Actions (V体育官网)
- Actions (VSports手机版)
- Actions (VSports注册入口)
- Actions (VSports app下载)
- "V体育2025版" Actions
- "V体育ios版" Actions
Substances
- Actions (VSports在线直播)
- Actions (V体育官网)
- "VSports最新版本" Actions
Grants and funding
LinkOut - more resources
"V体育安卓版" Full Text Sources
Other Literature Sources
VSports注册入口 - Medical
Molecular Biology Databases
"VSports app下载" Research Materials
V体育2025版 - Miscellaneous

