"V体育安卓版" The histone chaperone CAF-1 safeguards somatic cell identity
- PMID: 26659182
- PMCID: PMC4866648 (VSports在线直播)
- DOI: "VSports在线直播" 10.1038/nature15749
The histone chaperone CAF-1 safeguards somatic cell identity
Abstract (V体育2025版)
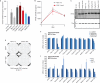
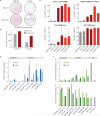
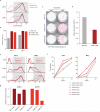
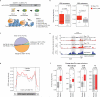
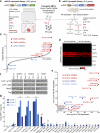
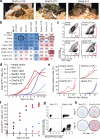
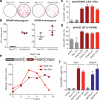
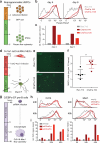
Cellular differentiation involves profound remodelling of chromatic landscapes, yet the mechanisms by which somatic cell identity is subsequently maintained remain incompletely understood. To further elucidate regulatory pathways that safeguard the somatic state, we performed two comprehensive RNA interference (RNAi) screens targeting chromatin factors during transcription-factor-mediated reprogramming of mouse fibroblasts to induced pluripotent stem cells (iPS cells). Subunits of the chromatin assembly factor-1 (CAF-1) complex, including Chaf1a and Chaf1b, emerged as the most prominent hits from both screens, followed by modulators of lysine sumoylation and heterochromatin maintenance. Optimal modulation of both CAF-1 and transcription factor levels increased reprogramming efficiency by several orders of magnitude and facilitated iPS cell formation in as little as 4 days VSports手机版. Mechanistically, CAF-1 suppression led to a more accessible chromatin structure at enhancer elements early during reprogramming. These changes were accompanied by a decrease in somatic heterochromatin domains, increased binding of Sox2 to pluripotency-specific targets and activation of associated genes. Notably, suppression of CAF-1 also enhanced the direct conversion of B cells into macrophages and fibroblasts into neurons. Together, our findings reveal the histone chaperone CAF-1 to be a novel regulator of somatic cell identity during transcription-factor-induced cell-fate transitions and provide a potential strategy to modulate cellular plasticity in a regenerative setting. .
Figures (V体育安卓版)





References
-
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi:10.1016/j.cell.2006.07.024. - PubMed
-
- Vierbuchen T, Wernig M. Molecular roadblocks for cellular reprogramming. Molecular cell. 2012;47:827–838. doi:10.1016/j.molcel.2012.09.008. - VSports最新版本 - PMC - PubMed
-
- Yang CS, Chang KY, Rana TM. Genome-wide functional analysis reveals factors needed at the transition steps of induced reprogramming. Cell reports. 2014;8:327–337. doi:10.1016/j.celrep.2014.07.002. - PMC (VSports app下载) - PubMed
Publication types
- Actions (V体育ios版)
- Actions (V体育平台登录)
- V体育ios版 - Actions
MeSH terms
- "V体育官网" Actions
- "VSports app下载" Actions
- "V体育官网入口" Actions
- "V体育官网" Actions
- "VSports app下载" Actions
- "VSports注册入口" Actions
- Actions (VSports在线直播)
- Actions (V体育安卓版)
Substances
- "VSports在线直播" Actions
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

