MicroRNA-31 initiates lung tumorigenesis and promotes mutant KRAS-driven lung cancer (VSports)
- PMID: 26657862
- PMCID: "VSports" PMC4701567
- DOI: 10.1172/JCI82720
V体育平台登录 - MicroRNA-31 initiates lung tumorigenesis and promotes mutant KRAS-driven lung cancer
Abstract
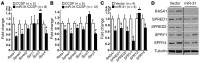
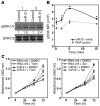
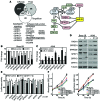
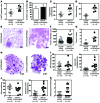
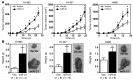
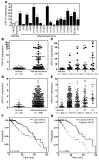
MicroRNA (miR) are important regulators of gene expression, and aberrant miR expression has been linked to oncogenesis; however, little is understood about their contribution to lung tumorigenesis. Here, we determined that miR-31 is overexpressed in human lung adenocarcinoma and this overexpression independently correlates with decreased patient survival. We developed a transgenic mouse model that allows for lung-specific expression of miR-31 to test the oncogenic potential of miR-31 in the lung VSports手机版. Using this model, we observed that miR-31 induction results in lung hyperplasia, followed by adenoma formation and later adenocarcinoma development. Moreover, induced expression of miR-31 in mice cooperated with mutant KRAS to accelerate lung tumorigenesis. We determined that miR-31 regulates lung epithelial cell growth and identified 6 negative regulators of RAS/MAPK signaling as direct targets of miR-31. Our study distinguishes miR-31 as a driver of lung tumorigenesis that promotes mutant KRAS-mediated oncogenesis and reveals that miR-31 directly targets and reduces expression of negative regulators of RAS/MAPK signaling. .
Figures




References
Publication types
- "VSports注册入口" Actions
MeSH terms
- "VSports注册入口" Actions
- Actions (V体育官网)
- Actions (VSports最新版本)
- V体育官网入口 - Actions
- VSports注册入口 - Actions
- Actions (VSports在线直播)
- "V体育平台登录" Actions
- VSports - Actions
Substances
- V体育ios版 - Actions
Grants and funding
- UL1 TR000445/TR/NCATS NIH HHS/United States
- UL1TR000445/TR/NCATS NIH HHS/United States
- R03 CA167695/CA/NCI NIH HHS/United States
- R01 CA177786/CA/NCI NIH HHS/United States
- R03CA167695/CA/NCI NIH HHS/United States
- T32 CA119925/CA/NCI NIH HHS/United States
- P50CA090949/CA/NCI NIH HHS/United States (V体育安卓版)
- R01 AR061474/AR/NIAMS NIH HHS/United States
- P50 CA090949/CA/NCI NIH HHS/United States
- P30 CA068485/CA/NCI NIH HHS/United States
- P30CA068485/CA/NCI NIH HHS/United States
- R01CA177786/CA/NCI NIH HHS/United States
LinkOut - more resources
"V体育官网" Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

